Teaming Up to Tackle Engineering Challenges
Innovation Awards Accelerate Development of New Research Techniques

This year’s Director’s Challenge Innovation Awards encouraged IRP collaborations between biomedical researchers and scientists with expertise in engineering or the physical sciences.
Scientists spend years, even decades, intensely studying a specific disease or biological system, an approach that yields unrivaled knowledge. However, many important scientific questions require a deep understanding of several subjects. As a result, the IRP has numerous programs dedicated to encouraging scientists with different areas of expertise to work together.
One such program is the NIH Director’s Challenge Innovation Awards, which funds innovative, high-impact projects that require the cooperation of researchers in more than one of NIH’s Institutes and Centers. This year, the program selected six promising proposals with one foot in the disciplines of biology and medicine and another in engineering or the physical sciences.
“Every round of the Innovation Awards has a different focus and is inspired by current areas of opportunity,” says Charles Dearolf, Ph.D., Director of Program Development and Support for the NIH’s Office of Intramural Research (OIR). “This round, the program wanted to support projects that will add to resources and collaborations within the growing bioengineering and physical sciences community at the NIH.”
By utilizing a variety of advanced technologies and drawing on expertise from several disciplines, the half-dozen projects chosen this year could lead to breakthroughs in our understanding of viral replication, our ability to detect tumors, and our knowledge of the brain, among other important innovations. Read on for a run-down of these efforts and their potential to transform the way we understand and treat disease.
Rapid Testing for a Dangerous Lung Infection
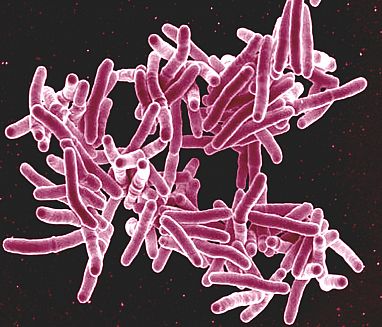
NIH’s Dr. Adrian Zelazny is attempting to develop a test for infections caused by close cousins of the bacterium that causes tuberculosis, pictured here.
Tuberculosis (TB), a lung disease caused by the bacterium Mycobacterium tuberculosis, was once one of the most feared illnesses in the world, killing one out of every seven people living in the U.S. and Europe in the late 1800s. Even today, the disease kills more than a million people per year globally. The large death toll from tuberculosis has long overshadowed infections caused by that microbe’s close cousins, a group of organisms known as non-tuberculous mycobacteria (NTM) that mostly affect people with weakened immune systems or existing lung problems.
“There are a lot of tools out there for diagnosing tuberculosis, but there are not as many for NTM,” says Adrian Zelazny, Ph.D., chief of the Microbiology Service in the NIH Clinical Center’s Department of Laboratory Medicine. “This is because, historically, people have just focused on TB and developed tools for diagnosing it, but those tools don’t work as well for non-TB mycobacteria.”
Dr. Zelazny has spent years working with Tulane University’s Tony Hu, Ph.D., on a test that uses the famed CRISPR system to diagnose TB infections. The test works by detecting Mycobacterium tuberculosis DNA in a person’s blood that is floating free outside of cells, a type of genetic material known as cell-free DNA. As a result, the test can return results much more quickly and detect smaller amounts of the bacteria than traditional methods of diagnosing TB.
Now, with the help of the Director’s Challenge Awards, Dr. Zelazny is launching an effort to adapt that test not just for detecting NTM, but also cell-free DNA with specific patterns of ‘epigenetic’ modifications that suggest the DNA originated in the lungs. The latter measurement can provide a readout of how severely NTM or another lung infection has damaged the organ.
“One of the things I like about this proposal is the combination of assessing both the pathogen and the patient’s clinical status, which I think is one of the failures of traditional diagnostic testing laboratories,” Dr. Zelazny says. “Testing tends to be compartmentalized by diagnostic interest and expertise, and the different parts may not necessarily communicate with each other. I like the idea of creating a test that combines the two sides. That’s the strength of this proposal: one test gives you both answers at the same time.”
Dr. Zelazny is collaborating on the project with IRP Lasker Clinical Research Scholar Sean Agbor-Enoh, M.D., Ph.D., whose own research at the National Heart, Lung, and Blood Institute (NHLBI) uses cell-free DNA to assess the health of donated organs received by heart and lung transplant patients. Once the CRISPR-based TB test has been re-engineered to detect cell-free DNA from NTM and the lungs, Dr. Zelazny will assess how well it works by applying it to blood samples collected via an existing study of NTM run by a research team led by IRP senior clinician Kenneth Olivier, M.D., M.P.H.
“I think it’s nice to get different Institutes to work together, and that is the intention of this type of award,” Dr. Zelazny says. “I had never worked with Sean before this, and actually I didn’t even know Sean before this. I was talking to Dr. Olivier about how it would be nice to add a human component to this test, and he referred me to Sean. That opened a new sort of direction I never thought I could do myself because I don’t have expertise in detecting lung damage. And now Sean is discussing his approach with Tony Hu, so it sort of creates this energy mass of people talking to each other who had never talked to each other before. That’s a nice thing to happen in science in general.”
Evaluating Toxins Using Synthetic Eyes

IRP researchers hope that using computer software to interpret video footage can make it easier to learn about the health effects of environmental chemicals.
The incredible intricacies of the human eye and brain allow us to see the world in stunning detail, but advances in artificial intelligence are increasingly giving similar abilities to computers. This so-called ‘machine vision’ uses complex programs to interpret video footage without the need for human involvement. While the technology is mostly associated with self-driving cars right now, it also has a huge number of other applications.
For instance, using funding from the Innovation Awards, Dondrae Coble, D.V.M, is leading an IRP effort to use machine vision to learn how chemicals found in the environment, such as pesticides and other pollutants, affect the behavior of fruit flies and mice. Since humans share many biological similarities with these species, substances that alter their behavior are likely to have some effect on us as well. However, determining how animals’ behavior changes after exposure to a chemical is far from easy.
“In addition to directly assessing nervous system dysfunction, which is the focus of ‘traditional’ behavioral assessments, behavioral change is an important physiological and translational endpoint for a broad spectrum of organ system diseases,” explains Dr. Coble, a veterinarian who leads the Comparative Medicine Branch at the National Institute of Environmental Health Sciences (NIEHS), as well as the Institute’s animal research program. “However, animal testing is the most time-intensive sort of testing, with behavioral tests being the most time- and labor-intensive, often costing millions of dollars for a single compound. Such continuous behavioral monitoring has not been feasible with traditional human observation due to the time and labor required.”
Developed via a collaboration between researchers at NIEHS, the National Institute of Biomedical Imaging and Bioengineering (NIBIB), and the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), the machine vision system Dr. Coble’s project relies on could eliminate the need for humans to review hours of footage in order to detect and meticulously document small changes in animal behavior. What’s more, the group plans to combine its system’s behavioral findings with information about how each chemical they test affects the activity of the animals’ genes. Establishing a relationship between changes in an animal’s behavior and how its genes behave could allow scientists to better predict how a chemical might affect other species, including ourselves.
“There is a growing push to replace animal studies with increasingly sophisticated cell-based model systems, but even the most complex of those novel systems significantly under-represents the complexity of the intact organism,” Dr. Coble says. “Accordingly, animal studies remain critical to our efforts to advance public health and mitigate human disease, though we must make every effort to refine these approaches to improve animal welfare and increase efficiency and translatability.”
Tracking Tweaks to Neuronal Superhighways
In many ways, cells are like tiny cities, complete with distinct structures that serve specific purposes and a complex system of biological roadways called microtubules that are used to transport life-sustaining materials from place to place. Microtubules are made up of a substance called tubulin, and just like street signs help vehicles navigate around cities, the cell can direct cellular traffic by modifying those tubulin building blocks in specific ways. This collection of tubulin tweaks, collectively called the ‘tubulin code,’ is essential for proper cellular health, especially in neurons, which need to move materials along their entire length, which can reach up to a meter.
“Microtubules are not inert tracks, but rather have information encoded in them through the tubulin code,” explains IRP senior investigator Antonina Roll-Mecak, Ph.D. “Perturbations in this code lead to early-onset neurodegeneration, so it is imperative that we understand how the tubulin code regulates cargo transport, microtubule dynamics, and organization in the neuron.”
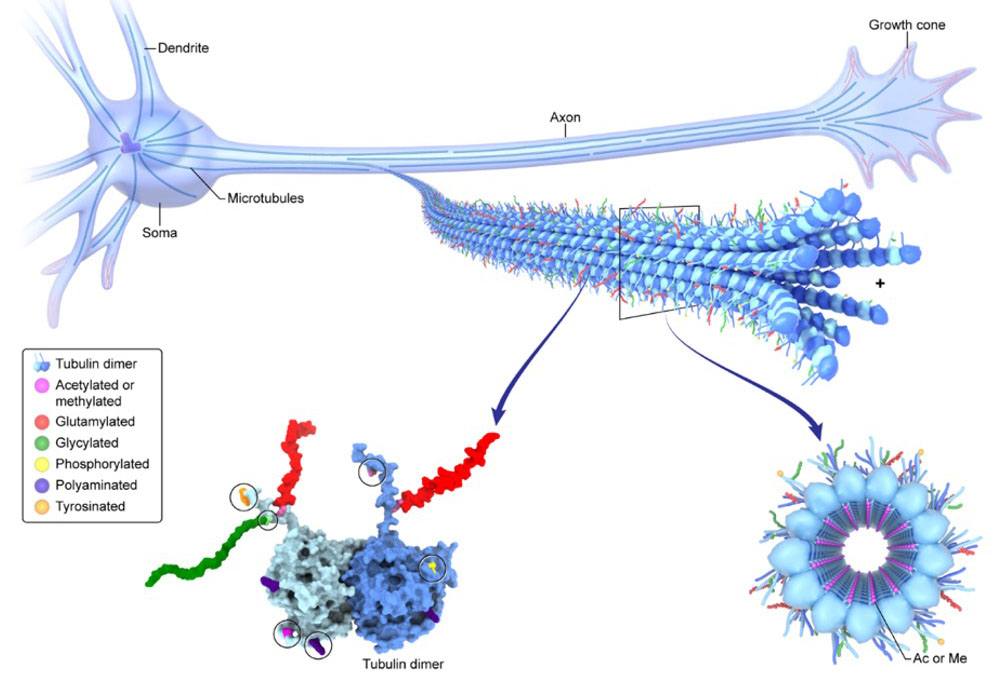
As neurons mature, they add different chemical modifications to the tubulin building blocks that make up their microtubules. Dr. Antonina Roll-Mecak wants to leverage high-resolution microscopes to record those changes over the course of a neuron’s lifespan.
Having spent her career studying tubulin, Dr. Roll-Mecak has made a great deal of progress toward cracking the tubulin code. Nevertheless, it is still unknown how the alterations nerve cells make to tubulin and the enzymes that carry out those adjustments change over time. To solve this mystery, Dr. Roll-Mecak’s lab is teaming up with the Advanced Imaging and Microscopy group at NIBIB to visualize in crisp detail how individual tubulin molecules are modified as neurons mature using the NIBIB team’s cutting-edge, high-resolution microscopes.
“This project is the perfect example of a synergistic collaboration,” Dr. Roll-Mecak says. “My lab has made fundamental discoveries in how the tubulin code regulates microtubule effectors, and the Advanced Imaging and Microscopy team brings unique, cutting-edge microscopy to this project. Together, we aim to bridge the resolution gap between my lab’s molecular discoveries and cellular physiology.”
“I am very excited about this award,” she continues. “It will allow us to answer fundamental questions about neuronal cell biology, while at the same time also making advances in cutting-edge imaging technologies that will be useful to other investigators in the field.”
Combining Pint-Size Antibodies to Improve Tumor Detection
The human immune system uses antibody molecules like the ones illustrated here to flag infectious threats. Dr. Ross Cheloha hopes the smaller, lab-designed antibodies his IRP team is developing could be used to improve cancer detection and treatment.
One of the main tools our bodies use to combat infections are antibodies, tiny molecular tags that adhere to invading bacteria and viruses and flag them for elimination by the immune system. Seeing how well this works, scientists like IRP Stadtman Investigator Ross Cheloha, Ph.D., have borrowed the concept for their own purposes.
“Antibodies are nature’s solution to generating molecules that bind specifically to any target of interest,” Dr. Cheloha explains. “In the past few decades, scientists and physicians interested in combating cancer recognized the special features of antibodies and have turned to harnessing their power to treat and characterize tumors.”
Researchers can design specialized antibodies in their labs that latch onto molecules found only on the surface of cancer cells, allowing clinicians to use them in combination with non-invasive imaging technologies to figure out where tumors are hiding out in the body. This provides critical information about whether a patient’s cancer has spread from its site of origin and whether treatment is successfully shrinking tumors. However, the bulky antibodies currently used for this purpose take a long time to circulate around the body, which slows down efforts to use them for cancer imaging. What’s more, since the antibodies are often attached to radioactive molecules that can be detected by imaging machines, the longer they are in the body, the more harm they might cause to our tissues.
To solve these problems, Dr. Cheloha’s team has created much smaller antibodies, called nanobodies, that can be labeled with a dye or radioactive molecule at a specific location. These miniature antibodies move around the body much more quickly than their full-size counterparts, enabling faster imaging with fewer potential side effects.
Using the funding provided by the Innovation Awards, Dr. Cheloha’s team will work with IRP cancer researchers led by Mitchell Ho, Ph.D., and Freddy Escorcia, M.D., Ph.D., to develop its nanobodies for use as cancer imaging agents, which can be done by attaching the labeled nanobody to a second nanobody that binds to tumors. Due to their smaller size, these cancer-targeting nanobodies could be particularly useful for detecting small collections of cancer cells that might otherwise be missed. They could also potentially be attached to cancer-killing chemicals and used not just for imaging but for treatment as well.
“The Director’s Challenge Innovation Award is both a huge honor and a major catalyst to spur new research directions,” Dr. Cheloha says. “I am a new investigator at NIH that opened my lab in the fall of 2020, so I’ve only been on NIH campus during ‘COVID times.’ One of the most difficult things about this from a research perspective is that the appropriate restriction on in-person meetings has stifled the informal conversations that often lead to new project ideas and collaborations. The existence of the Director’s Challenge Awards inspired our group of labs to come together to devise a project that makes use of each of our unique areas of expertise.”
Leveraging Robotics to Learn About Viruses
A cancer researcher and a virologist are teaming up to use robotics for probing the secret lives of viruses.
The best tools have applications beyond those intended by their creators, and the technologies developed by scientists are no exception. When IRP senior investigator Grégoire Altan-Bonnet, Ph.D., developed a robotic system to help his lab at the National Cancer Institute (NCI) gauge how immune cells respond to tumors, he may not have imagined that it would one day be used to learn about another of the immune system’s foes: viruses.
With the support of a Director’s Challenge Award, his lab is now working with a team at NHLBI led by his wife, IRP senior investigator Nihal Altan-Bonnet, Ph.D., to tweak that robotic system so it can be used to learn about how viruses interact with our cells. Normally, examining the dynamics of how numerous different viruses interact with a variety of cell types would take a large amount of time and effort. However, with a robotic helping hand and a sprinkle of ‘machine learning,’ the two researchers hope to more easily find out how quickly different viruses infect different cells, how the immune system fends them off, and how they enter, reproduce within, and exit from cells. Researchers can then use that information to identify potential targets for drugs meant to reduce the ability of viruses to replicate themselves within the body.
“Modeling the key modes of the viral lifecycle within host cells will offer mechanistic insights into the limiting steps for replication versus immune rejection,” Dr. Nihal Altan-Bonnet explains.
“We are very honored to receive this funding,” she adds, “and we are super excited to see what will come out of this project!”
Finding Needles in the Cellular Haystack
The human body contains a wide variety of cells from blood cells to lung cells to skin cells. To make things even more complicated, in some circumstances, not all the cells of a certain type are exactly the same. For example, in people with HIV who are receiving antiviral medications, the virus still hides out in a small subset of immune cells.
“They’re a tiny minority of all the cells that are present, but they’re the ones where the virus is,” explains IRP investigator Eli Boritz, M.D., Ph.D. “These cells are a barrier to HIV cure. A long-standing need in the field has been to be able to describe these cells in as detailed a way as possible.”
To learn more about these unique HIV-harboring cells, Dr. Boritz teamed up with bioengineering researchers at the University of California, San Francisco, to develop a technique called FIND-Seq. FIND-Seq enables HIV researchers like Dr. Boritz to specifically examine the activity of genes in the few cells that still harbor the virus in patients receiving treatment for the infection. However, Dr. Boritz suspected that many of his colleagues at NIH would also find the technology extremely useful, and not just for studying HIV.
“We completed our initial project with this technology, and it took a very long time, and after a tremendous amount of work, we made findings that, to me, were extremely exciting, so I knew there was going to be a need to do more of this in the HIV field,” he says. “I wanted to make the technology available here at NIH for us and other people who wanted to use it, and then try to maybe, for the purposes of making that a more robust effort, incorporate some other biological questions that might increase the demand and the sense of utility of the method.”
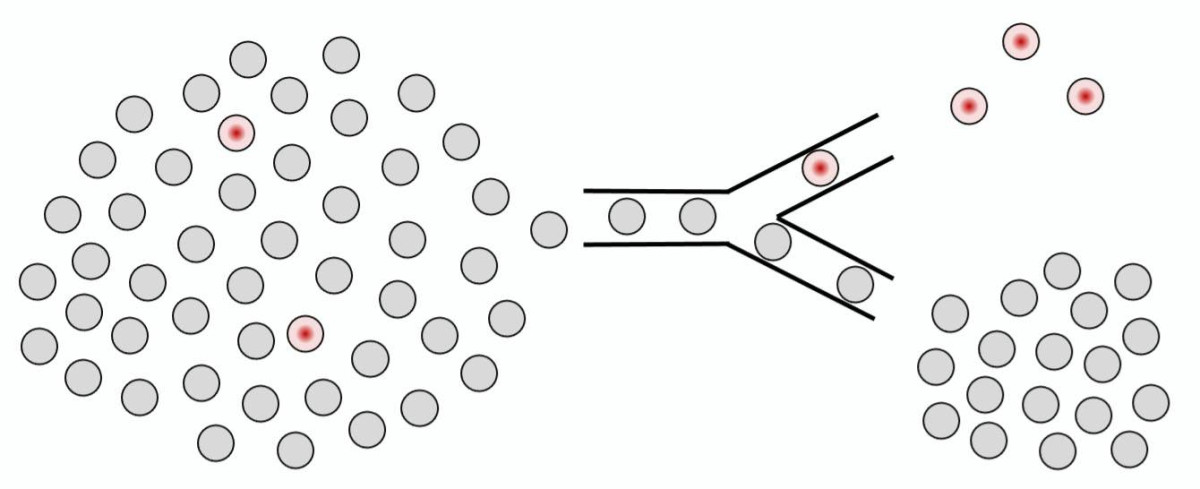
FIND-Seq can help scientists locate and study tiny subsets of cells that play outsized roles in viral infections and cancer.
Using funding from the Innovation Awards, Dr. Boritz aims to set up his FIND-Seq technology as a shared resource at NIH with the help of Nicole Morgan, Ph.D., who runs the Trans-NIH Shared Resource on Biomedical Engineering and Physical Science (BEPS) at NIBIB. He is also working with IRP senior investigator Christopher Hourigan, M.D., D.Phil., on adapting the technique so that it can be used to identify and study cancer cells in patients being treated for leukemia, a cancer that affects blood cells. Patients with leukemia often appear to be cancer-free after treatment only for their disease to come roaring back later on. This is thought to be because a tiny number of cancerous blood cells still remain in the body after treatment — too few to be studied using standard laboratory techniques, but perhaps not too few for FIND-Seq to sniff out. Using the technique to study how various genes behave in these cancerous holdouts could yield important insights into how to eliminate leukemia once and for all, just as Dr. Boritz hopes will eventually be the case for HIV.
“One thing that I learned as we were working on this project was that the more people who are thinking about it and using it and trying to apply it to their own research questions, the better it’s going to be,” Dr. Boritz says. “I think the level of the science goes up when there is trans-Institute collaboration and multiple biological questions under study and multiple research groups thinking about things. That has been very clear in my experiences all along.”
Subscribe to our weekly newsletter to stay up-to-date on the latest breakthroughs in the NIH Intramural Research Program.
Related Blog Posts
This page was last updated on Wednesday, May 24, 2023
