Research Briefs
Read about scientific advances and discoveries by NIH intramural scientists: biglycan protein found to be essential to bone repair; association between cannabis use and schizophrenia; genetic risk factors that contribute to dementia risk; new human pangenome captures genetic diversity; music-based intervention toolkit guides future studies; how omega-3 fatty acids get delivered to the brain.
NIDCR: SCIENTISTS FIND PROTEIN ESSENTIAL TO BONE REPAIR IN MICE
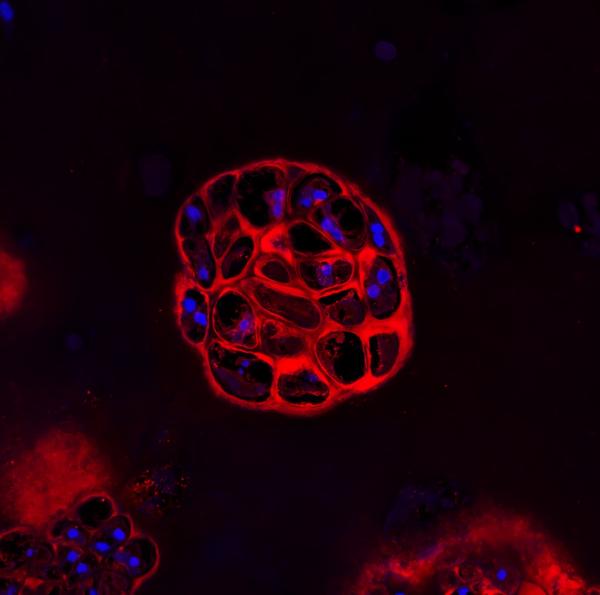
CREDIT: MARIAN YOUNG, NIDCR
Disrupting a sugar-laden protein called biglycan impaired bone growth and fracture healing in mice. Shown: Skeletal stem cells from healthy mice formed clusters of cartilage-producing cells, as shown, that were later replaced by bone.
A sugar-laden protein called biglycan appears central to bone integrity, strength, and proper fracture healing, according to an NIDCR-led study. The findings may help scientists understand why some fractures heal poorly and could provide insights into diseases marked by bone loss, such as osteoporosis.
To understand the role of biglycan in bone growth and healing, the scientists bred mice that lacked the protein. Compared with their healthy counterparts, mice born without biglycan had thinner, hollower, and weaker leg bones—signs resembling osteoporosis in humans.
The lack of biglycan also disrupted the normal sequence of fracture repair. In biglycan-deficient mice, fewer skeletal stem cells migrated to the fracture site to form a mass, or callus, that protects the fracture as it heals. Although the calluses eventually turned into new bone, the structure was irregular, indicating that the lack of biglycan can undermine bone healing.
A closer look at the mice’s skeletal stem cells hinted at a possible reason. During the normal healing process, skeletal stem cells first become cartilage-producing cells, which are later replaced by bone-forming cells. But stem cells isolated from biglycan-deficient mice and grown in the lab developed straight into bone, skipping the formation of cartilage, which normally serves as a template for new bone growth.
These findings suggest that biglycan’s role in bone healing may relate to its influence on skeletal stem cells. With more research, the study could inform strategies to better heal fractures and treat bone disorders. (NIH authors: R. Shainer, V. Kram, T.M. Kilts, L. Li, A.D. Doyle, D. Martin, and M.F. Young, Front Physiol 14, 2023; DOI:10.3389/fphys.2023.1119368)
[BY TIFFANY CHEN, NIDCR]
NIDA: ASSOCIATION BETWEEN CANNABIS AND SCHIZOPHRENIA IN YOUNG MALES
Researchers at NIDA and at the Copenhagen University Hospital of Denmark (member hospitals located in various cities) have found that young men with cannabis-use disorder (CUD) are at increased risk of developing schizophrenia.
In this study, the authors analyzed data from almost 7 million individuals in nationwide Danish health records over the course of 50 years. Researchers estimated the number of schizophrenia cases that could be linked to CUD at a population level.
Males with CUD in the 16–20 age group were more than twice as likely to develop schizophrenia compared with females. Similar results were shown in the 21–25 year age group. However, no differences were observed between the sexes for those aged over 25.
The authors estimated that 15% of schizophrenia cases among males aged 16–49 could have been avoided by preventing CUD in 2021. For young men aged 21–30, the number of preventable cases may be as high as 30%. In contrast, only 4% of schizophrenia cases in females aged 16–49 might have been prevented.
These findings highlight the importance of early detection and treatment of CUD, particularly in young males. “As access to potent cannabis products continues to expand, it is crucial that we also expand prevention, screening, and treatment for people who may experience mental illnesses associated with cannabis use,” said NIDA Director and study coauthor Nora Volkow. (NIH authors: W. Compton, E. Einstein, N.D. Volkow, and B. Han, Psychol Med 2023; DOI:10.1017/S0033291723000880)
[BY ANNELIESE NORRIS, NCI]
NINDS, NIA, NCATS: LARGE GENETIC CHANGES CONTRIBUTE TO DEMENTIA RISK
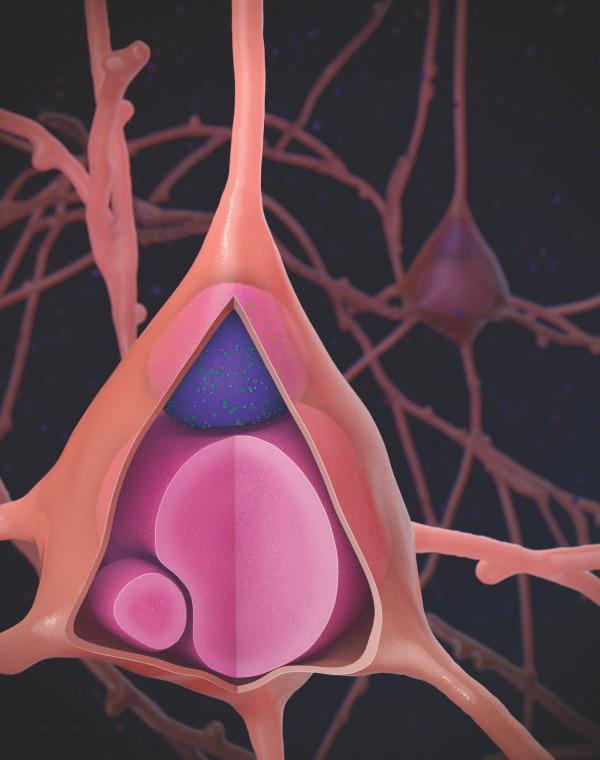
CREDIT: SONJA SCHOLZ, NINDS
Illustration showing a Lewy body (purple) within a neuron, the characteristic pathological feature of Lewy body dementia.
A study conducted by a trans-NIH group of investigators and their collaborators discovered several genetic risk factors for Lewy body dementia (LBD), frontotemporal dementia (FTD), and amyotrophic lateral sclerosis-associated (ALS) dementias. Non-Alzheimer’s dementias may account for up to 30% of all dementias in older adults.
The scientists analyzed thousands of DNA samples using computer algorithms and machine learning to identify structural variations (SVs). SVs are large regions of genomic variation that can represent hundreds, or even thousands, of nucleotides at once. In the FTD and ALS group, the researchers identified known SVs in the C9orf72 and MAPT genes, which demonstrated that the algorithms were working properly. In the LBD group, a previously unknown variant in the TPCN1 gene was discovered that is also associated with Alzheimer’s disease. The research team has made the analysis code and raw data available to all researchers to study their genes of interest.
Other rare pathogenic SVs likely associated with non-Alzheimer’s dementias were also revealed. “With each discovery, we shed light on the mechanisms behind neuronal cell death or dysfunction, paving the way for precision medicine to combat these debilitating and fatal disorders," said NIA Senior Investigator Bryan Traynor. (NIH authors: K. Kaivola, R. Chia, J. Ding, M. Rasheed, K. Billingsley, R. Dewan, A. Stark, A. Ray, S. Solaiman, P.A. Jerez, L. Malik, L. Ferrucci, S.M. Resnick, T. Tanaka, J. Raphael Gibbs, B.J. Traynor, and S.W. Scholz, Cell Genomics 3:100316, 2023; DOI:10.1016/j.xgen.2023.100316)
[BY STEPHEN ANDREWS, NCI]
NHGRI, NLM: NEW HUMAN PANGENOME REFERENCE CAPTURES GENOMIC DIVERSITY
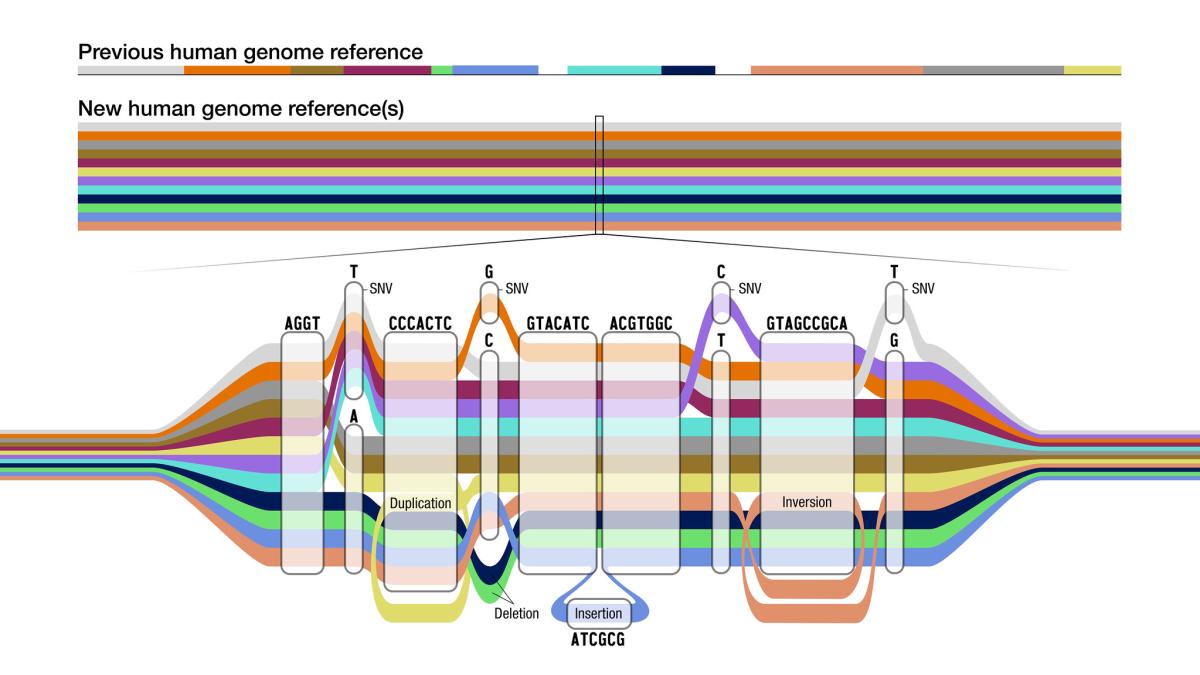
CREDIT: DARRYL LEJA, NHGRI
The new pangenome reference is a collection of different genomes from which to compare an individual genome sequence. Like a map of the subway system, the pangenome graph has many possible routes for a sequence to take, represented by the different colors.
NIH researchers and a team led by the NHGRI-funded international Human Pangenome Reference Consortium released a new draft human pangenome, incorporating sequences from 47 genetically distinct individuals from around the world.
Hailed as the “Book of Life,” the human reference genome released more than 20 years ago has been foundational in the field of human genetics. It originally consisted of genetic fragments from a small pool of individuals, and most of the sequence originated from one donor. Although refined through the decades, that reference does not reflect the vast genetic complexity of the human species, potentially biasing and introducing inequities in genomic analyses.
Most diversity within our species is driven by small genomic deviations. Those range from larger structural variants (SVs) to smaller single-nucleotide polymorphisms (SNPs), and insertions and deletions (indels). The updated pangenome more accurately identifies SNPs and indels that have important implications in the discovery and diagnosis of rare genetic disorders. Further, SVs, which have remained largely unexplored because of the limitations of using a single reference genome, are more easily identified, broadening the horizons of future genome-wide association studies which detect genetic markers of disease.
Future work will expand this resource to include full-genome analyses of at least 350 individuals within a diverse global cohort, ultimately bringing more equitable health information to people around the world. (NIH authors: A.L. Felsenfeld, V.A. Schneider, B.I. Schultz, M.W. Smith, H.J. Sofia, F. Thibaud-Nissen, S. Koren, A. McCartney, S. Nurk, M. Rautiainen, A. Rhie, B. Walenz, and A.M. Phillippy, Nature 617:312-324, 2023)
[BY TAYLOR FARLEY, NIAID]
NCCIH, NIA, NINDS, NIDCD, NHGRI: NIH SCIENTISTS CREATE MUSIC-BASED INTERVENTION TOOLKIT
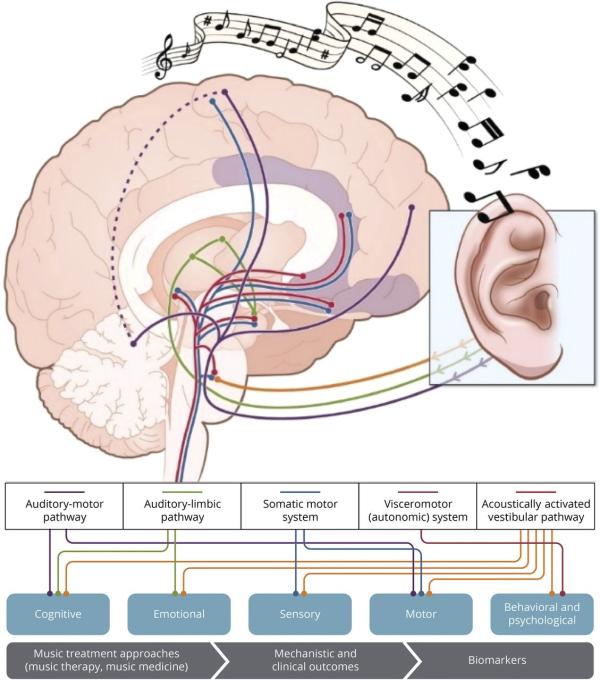
CREDIT: ADAPTED WITH PERMISSION FROM NAT REV NEUROSCI 15:170-180, 2014
Illustration showing the pathways underlying the neural and physiologic responses to music.
For neurologic disorders, such as stroke and traumatic brain injury, music-based interventions (MBIs) have shown tantalizing potential to improve quality of life by helping to manage symptoms, slow disease progression, and facilitate rehabilitation. Patients may prefer this approach because it can be affordable, accessible, and noninvasive. Yet, precisely how MBIs should be applied to therapy has not been established; the evidence has been anecdotal, limited to small-scale clinical trials or not designed within a scientific framework.
Enter the NIH MBI Toolkit, a resource created by representatives from the Trans-NIH Music and Health Working Group, the Renée Fleming Foundation, the Foundation for the NIH, and other experts and collaborators. The toolkit is intended to guide future research on music and human health and to provide standards and tools for investigators seeking NIH funding for MBI studies.
The NIH MBI Toolkit provides guiding principles and implementation strategies to design high-quality experiments and drive reproducible results: Essential elements to investigate, consistent descriptive terminology to differentiate among different types of MBIs, and relevant biomarkers to assess treatment response. The toolkit also highlights proven assessment methods that allow collection of rigorous and robust behavioral measurements such as alertness, language, memory, and motivation.
Although the authors focused on brain disorders of aging, they envision that the MBI Toolkit might also be modified for use in other diseases across the lifespan. (NIH authors: E. Edwards, C. St. Hillaire-Clarke, D.W. Frankowski, R. Finkelstein, T. Cheever, W.G. Chen, L. Onken, A. Poremba, R. Riddle, D. Schloesser, C.E. Burgdorf, N. Wells, and F.S. Collins, Neurology 100:868-878, 2023; DOI:10.1212/WNL.0000000000206797)
[BY SEPPIDEH SAMI, CC]
NICHD: ZEBRAFISH MODEL REVEALS HOW BRAIN ACQUIRES OMEGA-3 FATTY ACIDS
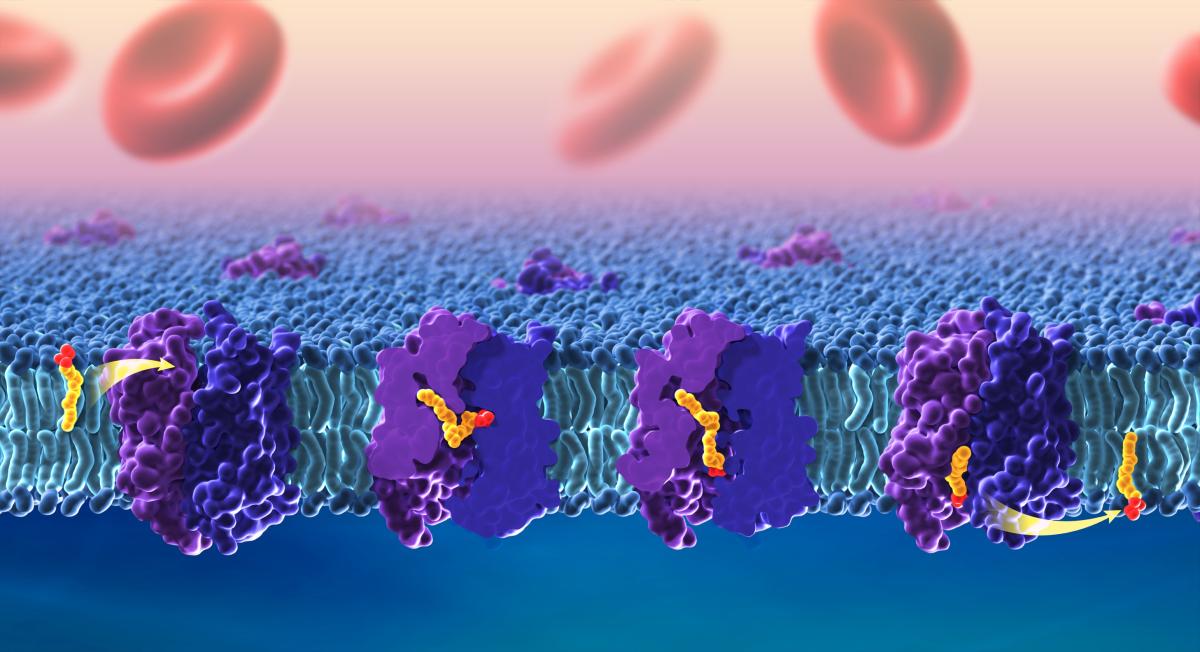
CREDIT: ETHAN TYLER, NIH MEDICAL ARTS
Docosahexaenoic acid (DHA) is an essential omega-3 fatty acid that is important for a healthy nervous system. This model shows how DHA and other omega-3 fatty acids cross the blood–brain barrier through the lipid transporter Mfsd2a.
Omega-3 fatty acids are vital to the development of the brain, where they act as a primary structural component of cell membranes. Sourced solely through our diet, omega-3 fatty acids must cross the blood–brain barrier by interacting with selective transporters called flippases. One of the most abundant of those is Mfsd2a, and mutations in this protein have been linked to neurological conditions such as dyspraxia, dyslexia, and microcephaly. Using a zebrafish model (Danio rerio), NICHD researchers and their colleagues are among the first to show precisely how Mfsd2a delivers omega-3 fatty acids to the brain.
By using single-particle cryoelectron microscopy, the investigators took snapshots of the Mfsd2a protein as it flipped the omega-3 fatty acid in the form of DHA-LPC across a cell membrane. The resulting images and subsequent 3D structural determination of Mfsd2a revealed the most detailed molecular analysis of Mfsd2a to date and identified three pockets within the protein used to maneuver fatty acids, in contrast to a previously proposed linear tunnel model.
The findings shed light on the mechanisms of other similar fatty acid transporters and could help inform how drugs are delivered across the blood–brain barrier. (NIH authors: L.T.F. Lai and D. Matthies, Nat Commun 14:2571, 2023; DOI:10.1038/s41467-023-37702-7)
[BY JONATHAN CHU, NIAID]
This page was last updated on Saturday, January 18, 2025
