The Virus vs the Machine
IRP Leverages Supercomputing to Combat Coronavirus

With 107,000 processing cores, the NIH's Biowulf supercomputer lends tremendous computational power to IRP efforts to curb the COVID-19 pandemic.
Over the past six months, a tiny virus has completely upended life in the United States and many other countries. To combat this microscopic threat, some IRP researchers have turned to a tool the size of a small building.
Biowulf, the NIH’s supercomputer, is supporting more than a dozen different IRP research projects focused on the novel coronavirus. As the world’s most powerful supercomputer solely dedicated to biomedical research, Biowulf allows scientists to analyze data and run simulations at unprecedented speed. Two weeks ago, a blog post described how IRP investigators are using Biowulf to elucidate the structure of the novel coronavirus and simulate how potential therapeutics might interact with it. Picking up where that post left off, this blog will explore the application of Biowulf to important questions about the spread of COVID-19 and the way that its genes, along with our own, might influence its impact on the body.
Delving Into Genomic Data
The novel coronavirus may have a long genome as far as viruses go, but its 30,000 base pairs don’t hold a candle to the approximately three billion base pairs that make up the human genome. It should come as no surprise, then, that several research groups are leveraging Biowulf’s computational might to answer questions about how COVID-19 affects our genes, and vice-versa.
A consortium of researchers directed by IRP senior investigator Helen Su, M.D., Ph.D., for example, is using Biowulf to help sequence and analyze the genomes of previously healthy patients who experienced a severe or fatal COVID-19 infection. The group is specifically searching for genetic variants that reduce an individual’s ability to fight off the virus. Once they have identified potentially influential mutations, the IRP researchers will perform laboratory experiments to determine whether the genetic variants impair cells’ capacity to respond to infection by the novel coronavirus.

Several groups of IRP researchers are examining our genes and their behavior for clues as to why some COVID-19 patients experience more severe symptoms than others.
Meanwhile, scientists in the lab of IRP senior investigator Lothar Hennighausen, Ph.D., are utilizing NIH’s supercomputer to examine how COVID-19 affects the activity of genes in disease-fighting white blood cells called leukocytes. Using data from roughly 400 infected and non-infected individuals who were connected to a COVID-19 outbreak at a ski resort in Ischgl, Austria, Dr. Hennighausen’s team and his Austrian collaborators hope to connect differences in the behavior of genes in these immune cells to differences in the patients’ symptoms, as well as whether or not they developed lingering complications that persisted after they recovered from COVID-19. Similarly, researchers led by IRP senior investigator Robert Balaban, Ph.D., and Mehdi Pirooznia, M.D., Ph.D., director of the Bioinformatics and Computational Core Facility at the National Heart, Lung, and Blood Institute (NHLBI), are using Biowulf to analyze the behavior of genes in human lung cells infected with the novel coronavirus and comparing it to the activity of genes in healthy cells and cells infected with other viruses, such as the related coronavirus that causes Middle East Respiratory Syndrome (MERS) and a specific form of the flu virus. Examining these changes in gene activity will permit the IRP team to identify cellular processes that may be uniquely affected by COVID-19, thereby providing targets for therapeutics.
“Next-generation sequencing (NGS) technology relies heavily on having enough computing power and storage to perform large scale genomic processing,” Dr. Pirooznia says. “The latest NGS sequencers now routinely produce several gigabytes to terabytes of data. Here at the NIH, Biowulf is ideally designed to sort through the massive datasets required for computational genomics.”
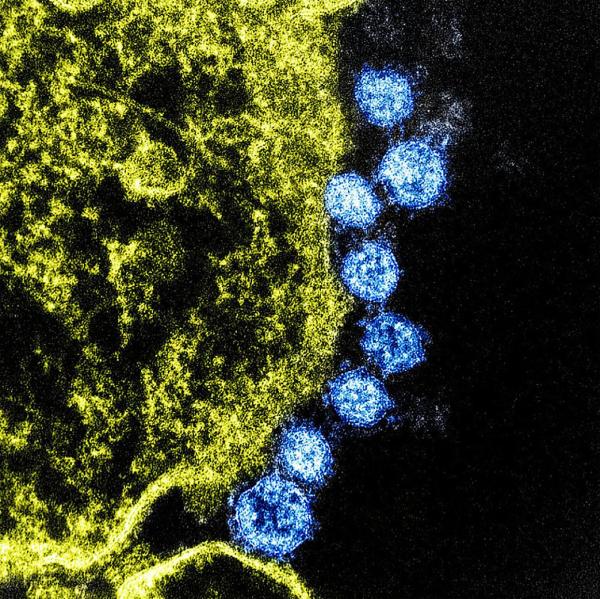
Viruses (blue) hijack the process of metabolism in infected cells (yellow) in order to fuel their own reproduction. Therapeutics that prevent that could help speed the recovery of COVID-19 patients.
The lab of IRP senior investigator Eytan Ruppin, M.D., Ph.D., is exploring the virus’ effects on gene activity as well, but his team’s focus is on cellular metabolism, the set of chemical processes that cells use to keep themselves alive, including converting food into energy. Because viruses need host cells to reproduce, they must redirect the metabolism of an infected cell so that it manufactures molecules that the virus requires rather than what the cell itself needs. Consequently, preventing the virus from hijacking metabolism in infected patients’ cells could hinder its reproduction. In collaboration with the lab of Sumit Chanda, Ph.D., at Sanford Burnham Prebys Medical Discovery Institute in La Jolla, California, Dr. Ruppin’s team — led by Kuoyan Cheng, a research fellow in Dr. Ruppin’s lab — is feeding data on the activity of metabolism-related genes into Biowulf to enable the supercomputer to simulate how COVID-19 might alter cellular metabolism and determine whether certain drugs might hinder replication of the virus by counteracting these changes. Currently, Dr. Ruppin’s lab is focusing its efforts on identifying drugs that can be combined with the already approved COVID-19 treatment remdesivir to increase its effectiveness.
“Our analysis involves solving thousands of mathematical optimization problems, which require very extensive computational resources,” Dr. Ruppin says. “Thus, a high-performance computing resource like Biowulf is indispensable for a project of this kind, and we are very grateful for all the help with got from the Biowulf team throughout the course of this project.”

Certain species of bats are among the variety of organisms that serve as natural reservoirs for coronaviruses like the one that causes COVID-19. Image credit: Anton Croos via Wikimedia Commons
Finally, IRP investigator Xiaofang Jiang, Ph.D., hopes delving into the novel coronavirus’ past will help public health officials prevent transmission of COVID-19 in the future. By utilizing Biowulf to mine publicly available collections of viral genomic data, her lab aims to identify the ‘reservoirs’ of coronaviruses — that is, the organisms that naturally carry the virus responsible for COVID-19, as well as its close cousins. In addition, Dr. Jiang’s team is comparing the genomes of the novel coronavirus with those of other coronaviruses in order to shed light on its evolutionary history, including specific events in which it swapped genes with other viruses. Learning more about how the virus evolved and jumped from animals into humans could help prevent future outbreaks of the novel coronavirus and other, similar viruses.
Simulating the Spread of COVID-19
A supercomputer like Biowulf can simulate not only what might happen when two molecules meet or how a virus might alter the metabolism of a cell, but also how a disease spreads from person to person. This sort of ‘epidemic model’ uses epidemiological data such as positive and negative test results, hospitalizations, and deaths to inform our understanding of how a disease is transmitted through a population and the range of possible scenarios that might unfold as an outbreak progresses. Supercomputers can simulate the outcomes of huge numbers of models, each slightly different from one another, and compare the simulations to real-world data in order to determine how closely each model’s results match the data and therefore how useful each model might be for predicting future conditions.
“There is a huge space of possibilities for how a model could be configured to give rise to the real-world data,” says Jonathan Fintzi, Ph.D., a statistician in the lab of IRP senior investigator Dean Follman, Ph.D. “The vast computing resources available to us at the NIH are critical to our ability to fit and evaluate such models.”
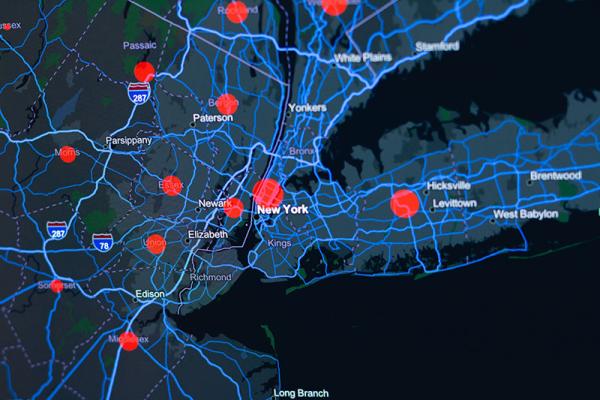
Supercomputers like Biowulf are able to simulate the spread of diseases in specific locations and under certain conditions, providing information that can help public health officials curb transmission. Image credit: Center for Systems Science and Engineering (CSSE) at Johns Hopkins University
Dr. Fintzi is using Biowulf to model the spread of COVID-19 at the state and local level, an effort that could yield insights into some of the variables that influence the virus’ transmission and assist with predictions about its future prevalence in specific locations. That information could, in turn, help public health officials design strategies to curb transmission of the disease. These efforts could also benefit from the results of research led by IRP senior investigator Carson Chow, Ph.D., who is using Biowulf to crunch epidemiological data in order to estimate how many individuals infected with COVID-19 are never identified and therefore not included in official case counts. His work could be particularly helpful for determining how many people have caught the virus and how often the disease is fatal.
Finally, IRP senior investigator Vipul Periwal, Ph.D., along with two postdoctoral fellows in his lab, Jungmin Han, Ph.D., and Evan Cresswell-Clay, Ph.D., used Biowulf to evaluate more than 56,000 models of COVID-19’s transmission using data confined to the period before measures were put in place to mitigate the virus’ spread. The study, which has yet to be published, found a 1.7 percent fatality rate for the disease and estimated that infected individuals are contagious, on average, for 22 days, though this could range from as few as 16 days to as long as 28 days. The IRP team's analysis also confirmed the effectiveness of social distancing measures, as even minimal social distancing dramatically reduced the number of infections in its simulations.
Without a supercomputer, evaluating thousands of mathematical models and sequencing billions of DNA base pairs would take an enormous amount of time. As COVID-19 continues to spread rapidly in many countries, any technology that can speed up the expansion of our knowledge about the disease will prove enormously beneficial. With the help of Biowulf and its dedicated staff, IRP researchers can rapidly make discoveries that will be crucial to stemming the tide of the coronavirus pandemic.
Subscribe to our weekly newsletter to stay up-to-date on the latest breakthroughs about the novel coronavirus and many other diseases from the NIH Intramural Research Program, and check out our previous blog post to learn more about how IRP scientists are using Biowulf to study COVID-19.
Related Blog Posts
This page was last updated on Wednesday, May 24, 2023
