IRP’s Eugene Koonin Elected to National Academy of Medicine
Scientist Decoded DNA to Build a Genomic Tree of Life

Dr. Eugene Koonin was elected to the National Academy of Medicine in October for his pioneering efforts to identify important DNA sequences shared by many different species.
In 1973, the geneticist Theodosius Dobzhansky wrote a now-famous essay that declared, “Nothing in biology makes sense except in the light of evolution.” That sentiment has served as the guiding principle for the career of IRP senior investigator Eugene V. Koonin, Ph.D., who was elected to the National Academy of Medicine (NAM) in October 2022 for his contributions to the field of evolutionary biology.
Dr. Koonin’s pioneering efforts to identify clusters of similar genes found in different organisms passed down by a common ancestor — known as ‘homologous’ genes — has helped to unlock the secrets encoded in DNA and create a foundation for the systematic study of how genes evolve and function. His lab at the National Library of Medicine’s National Center for Biotechnology Information (NCBI) uses a combination of genomic sequencing and mathematical modeling to compare genes across species and determine how they work and where they came from. From this information, his team can develop a systematic framework to show the relationship between genes as they evolved. It’s like drawing the tree of life, but on a genomic scale.
“My kind of computational biology, evolutionary genomics, stems from this necessity to understand what is written in those DNA sequences,” Dr. Koonin explains. “What is encoded there is essentially all the properties of organisms, including, of course, information related to health and disease. DNA tells us what we are, where we are similar to each other in basic features and where we are different, and why some of us are healthy and some less so.”
Thanks to the improvement of DNA sequencing technology over the past few decades, scientists have amassed an enormous amount of genomic data about all kinds of organisms, from tiny, genetically simple viruses to animals and humans. However, all those strings of A’s, T’s, C’s, and G’s that make up DNA are useless unless we can read them. The key to breaking the code is to use high-powered computers to compare genomes across species, which enables researchers like Dr. Koonin to identify ‘conserved’ genetic sequences that appear in many different organisms, making them likely candidates for genes that produce important traits or govern essential biological processes. In this way, the efforts of Dr. Koonin and other evolutionary biologists produce a kind of genetic Rosetta Stone that allows us to interpret the language of life and read its messages.
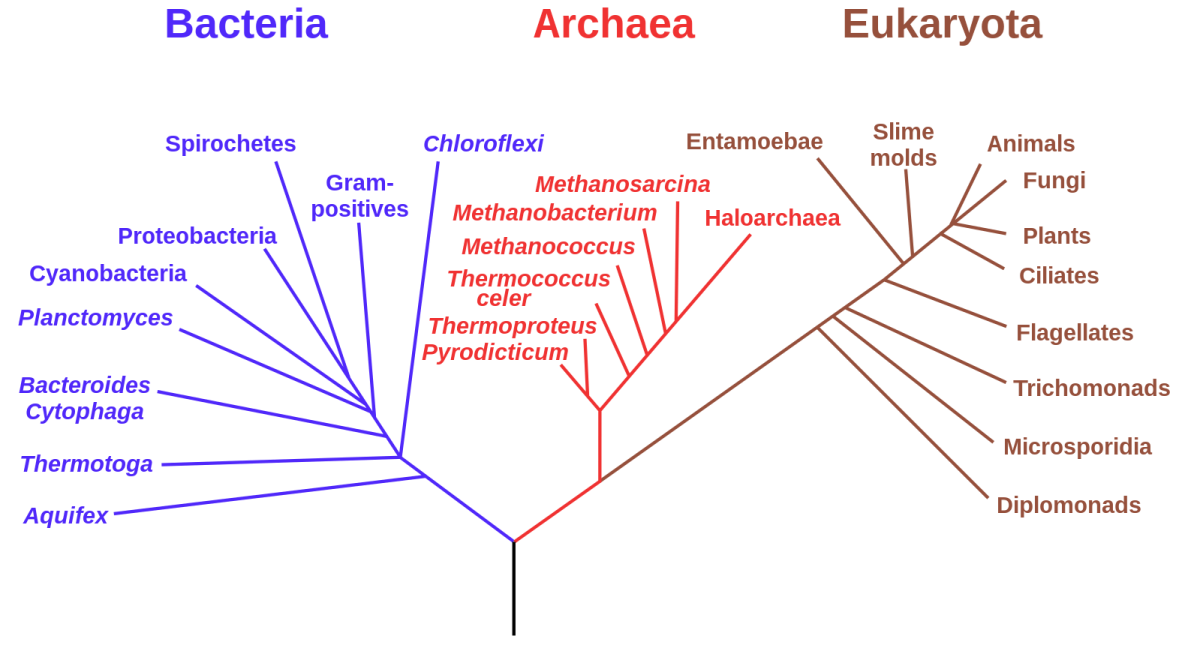
So-called ‘phylogenetic’ tree of life diagrams like this one show how very different organisms descended from a common evolutionary ancestor. As a result of this process, many distantly related species nevertheless have identical or similar genes in their genomes.
“When you know a sequence of DNA codes for a certain protein, this gives you a lot of predictive power,” Dr. Koonin says. “You can look at other genome sequences, sometimes even very distantly related ones, and find a related sequence in order to develop a testable hypothesis on the function of the protein and the amino acids that are essential for that function.”
Dr. Koonin’s interest in conserved DNA sequences isn’t limited to those that produce proteins. For example, he has long theorized that that ‘spacer’ DNA that does not code for proteins might play a role in the evolution of bacteria’s defense systems against viruses that prey on them. Discoveries from studies examining this idea by Dr. Koonin and others turned out to be a major step in developing the revolutionary genetic editing technique known as CRISPR/Cas9, which leverages the way bacteria remember and chop up the DNA of viruses that attack them.
One of the most important practical applications of Dr. Koonin’s research has been the development of the revolutionary gene editing technique called CRISPR/Cas9, which is based on insights into how bacteria chop up the DNA of viruses that attack them.
While CRISPR/Cas9 has a ways to go before it is routinely used in medical settings, the ability to classify genetic sequences based on their evolutionary function has already aided significant advances in medicine, especially in our ability to predict and protect ourselves from new threats. For example, Dr. Koonin’s lab developed a system to map the evolution of viruses and categorize them based on the characteristics they share. This system became the foundation for the global taxonomy of viruses used around the world to classify and name viruses, such as the noroviruses responsible for the ‘stomach flu’ and coronaviruses like the one that causes COVID-19. Importantly, Dr. Koonin’s research has shown that new viruses discovered nowadays continue to fit into the divisions his team defined years ago, making it easier to work out how a particular virus evolved, where it came from, how it replicates, and what might happen when humans become infected with it.
“When a new virus is discovered, particularly one that causes human disease, it’s important to be able to quickly determine what other viruses it is related to and, accordingly, what the different parts of its genome do,” Dr. Koonin says. “How can the virus change? And what can we possibly do to interfere with it?”
Dr. Koonin’s discoveries and classification frameworks have helped us better understand our genomic world, creating a foundation to answer important questions in health and revolutionizing both biological research and modern medicine. Today, he continues to build on his past accomplishments by developing methods to support the discovery of new antiviral defenses in bacteria, including discovering new CRISPR/Cas systems in bacteria that might be useful for gene editing. He is also exploring the DNA of cancer cells because the genetic changes that give rise to tumors resembles the process of how whole organisms evolve.
“Being elected to the National Academy of Medicine has special significance to me because my own research is primarily in fundamental areas of evolutionary biology,” Dr. Koonin says. “So, this is a recognition of the relevance of my work for human health.”
Subscribe to our weekly newsletter to stay up-to-date on the latest breakthroughs in the NIH Intramural Research Program.
Related Blog Posts
This page was last updated on Wednesday, July 5, 2023
