IRP Pushes Forward in Fight Against Pandemic Virus
Many NIH Labs Remain Focused on COVID Research
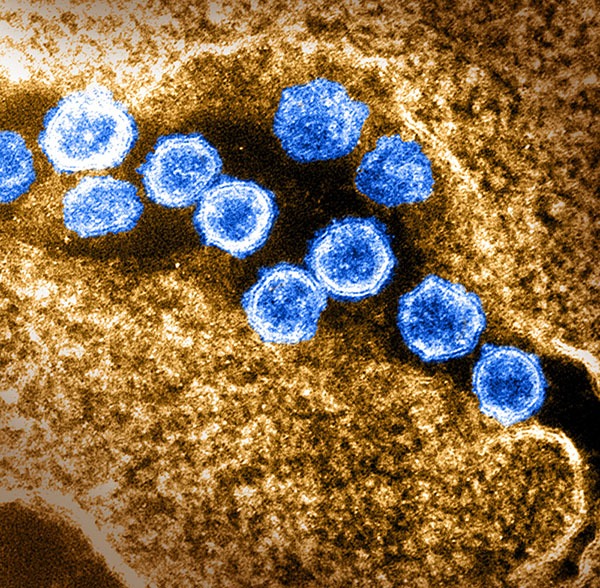
As the novel coronavirus responsible for the COVID-19 pandemic (blue) continues to spread widely, IRP researchers remain hard at work developing new treatments and methods to prevent infections.
Since the early days of the COVID-19 pandemic, IRP researchers have been hard at work learning about the virus and developing therapies for it. That research remains as important as ever, particularly as the new Omicron variant of the virus continues to spread rapidly.
Fortunately, NIH’s Intramural Targeted Anti-COVID-19 Program (ITAC) has been providing IRP scientists with millions of dollars to support their research on the pandemic virus, known as SARS-CoV-2. Last week, the “I Am Intramural” blog discussed ITAC-funded efforts to learn about the biology of the virus and how it affects the body. This week, we’ll look at IRP projects focused on ways to track, treat, and prevent infections.
Developing Drugs to Disrupt COVID-19
The current generation of COVID-19 vaccines offer fantastic protection against severe illness and death, but they are not perfect, and some vaccinated individuals — particularly those with weakened immune systems and other pre-existing conditions — will still come down with serious cases of the illness. Many more vaccinated people will become infected and experience non-life-threatening symptoms that are nevertheless still no fun to contend with. As a result, many IRP scientists are using ITAC funding to create new treatments for COVID-19.
In his own efforts, IRP senior investigator Mitchell Ho, Ph.D., is taking inspiration from an unexpected source: camels. Our own bodies produce molecules called antibodies that help combat infections, and they have four parts that bind to foreign materials in the body, such as viruses and bacteria. On the other hand, certain animals make smaller antibodies that have only two regions that bind to the antibody’s target. By isolating these two binding regions, scientists can create so-called ‘nanobodies’ that are small enough to attach to parts of an invading pathogen that our larger antibodies cannot. Nanobody-based therapies also wouldn’t require patients to come to the hospital for infusions like the antiviral drug remdesivir or the monoclonal antibodies currently being used to treat COVID-19. Finally, they are easier to manufacture than other antibody-based therapies.
“They can be easily produced in E. coli at much lower cost compared to conventional antibody drugs that are normally made in mammalian cells,” Dr. Ho explains. “Nanobodies are also great building blocks for making multivalent and multi-specific drugs, and they are highly soluble and stable, so they can be engineered as inhaled drugs for treating COVID-19 and other respiratory diseases.”
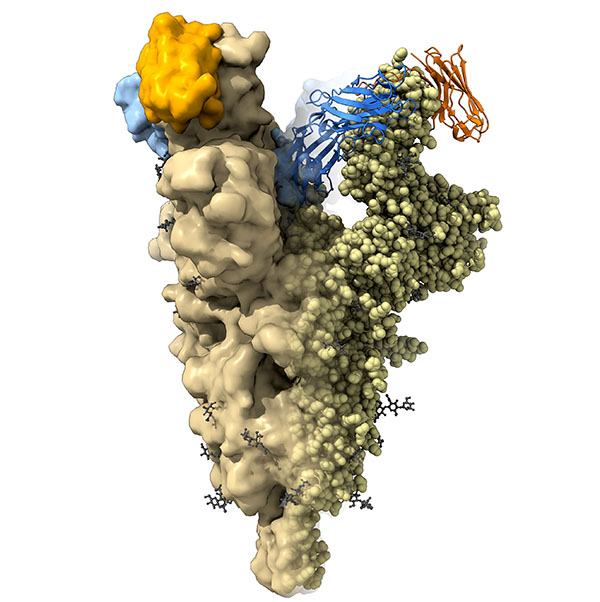
This structural image shows camel nanobodies (blue and orange) bound to the spike protein of SARS-CoV-2 to neutralize the virus.
Dr. Ho’s lab has spent months testing a large number of nanobodies created using camel antibodies for their effects on SARS-CoV-2 and its variants, including the Delta variant. So far, his lab has identified two nanobodies that bind tightly to parts of the novel coronavirus’ spike protein that are crucial for successful infection. Moreover, mouse studies have shown that one of these nanobodies protects mice from dying when infected with two different variants of the virus. Since the two nanobodies target different parts of the spike protein, Dr. Ho’s team has also been examining the therapeutic effects of combining them.
An IRP team led by NIH Distinguished Investigator T. Jake Liang, M.D., has also undertaken an effort to find new treatments for COVID-19, making use of its expertise on antiviral development for hepatitis viruses. Prior to the emergence of the novel coronavirus, his lab was working on several molecules that could stop the hepatitis C virus from infecting cells. Since there are commonalities between how hepatitis C and SARS-CoV-2 trespass into our cells, Dr. Liang’s team has been testing several of its hepatitis C inhibitors as treatments for COVID-19.
Two molecules in particular have shown promise at stopping several variants of the novel coronavirus from infecting cells in petri dishes, including the Delta variant. Moreover, his lab’s studies suggest that rather than interfering with the virus’ ability to attach to cells, the two molecules block a later step in infection during which the virus fuses its membrane with that of the cell it is attempting to hijack. While Dr. Liang believes the two molecules could be packaged into pills that COVID-19 patients could take at home, his team continues to search for and test additional compounds that could be even more effective against the disease.
“We don’t know whether these two compounds will be the leads that we hope to take into the clinic,” Dr. Liang says. “We are developing more active and pharmacologically suitable derivatives of these compounds that may be better for clinical development.”
Blocking a virus from infecting cells is not the only way to fight it. Other researchers hope to incapacitate the virus before it can even attempt to infiltrate our cells. Chemical disinfectants like soap and hand sanitizer do this when we apply them to our hands, but in addition to tasting awful, they would be harmful to ingest. Instead, researchers led by IRP senior investigator Alan T. Remaley, M.D., Ph.D., are developing new compounds that are ‘amphipathic,’ meaning one part of them is attracted to water molecules and the other is not. Amphipathic compounds can bind to molecules called lipids, which are found in the fat and cholesterol that naturally occur in our bodies but also in the cell membranes of bacteria and the envelopes that enclose virus’ genetic material. Prior to the pandemic, Dr. Remaley’s lab was working on treatments for cardiovascular disease, including one meant to mimic a natural molecule called ApoA-I that helps move cholesterol out of our cells.
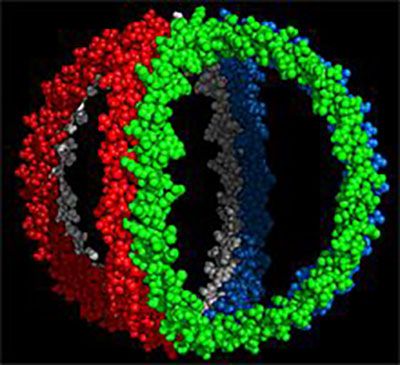
Image credit: W. Sean Davidson, University of Cincinnati
ApoA-I molecules — each a different color — form a cage-like structure that surrounds cholesterol-transporting molecules known as lipoproteins. Dr. Alan T. Remaley’s lab is leveraging their knowledge of ApoA-I to create similar molecules that might inactivate the SARS-CoV-2 virus.
“ApoA-I is a tandem array of amphipathic helices, which enables it to bind to lipids and remove cholesterol,” Dr. Remaley explains. “We used our expertise in this area to design new amphipathic peptides that bind to lipids on the viral envelope of the SARS-CoV2 virus.”
Dr. Remaley’s lab has so far designed and synthesized 33 amphipathic molecules and examined their ability to stop a virus belonging to the same family as SARS-CoV-2 from infecting cells in the lab. Some of the molecules did inactivate the virus, suggesting that Dr. Remaley’s approach could work, but much more will need to be done to come up with a drug that inactivates the novel coronavirus without harming our own cells. Moving forward, his lab’s efforts will focus on refining three compounds that show the most promise and, with the help of other intramural researchers, testing those compounds’ ability to neutralize the actual SARS-CoV-2 virus in a facility with the proper safety procedures to conduct such experiments.
New Ways to Test COVID Treatments
Indeed, as important as it is to identify and develop treatments for COVID-19, this work would all be for nothing without the ability to test them. Fortunately, many scientists are coming up with ways to gauge the ability of both new and existing drugs to interfere with the novel coronavirus. To that end, researchers in NIH’s National Center for Advancing Translational Sciences (NCATS) have designed more than a dozen highly sensitive experimental methods for determining how well a compound binds to or interferes with either viral molecules or human proteins that the virus requires for its life cycle, as well as assessing the effects of these interactions. These tests are being utilized in a ‘high-throughput screening’ process to rapidly gauge the potential of thousands of different compounds as treatments for COVID-19.
What’s more, a team of researchers led by IRP scientific project manager Kyle Brimacombe developed a website that provides real-time access to the results of NCATS’ drug-screening efforts. Using the online resource, called the NCATS OpenData Portal, scientists both within and outside NIH, as well as members of the public, can easily view the results of those experiments, enabling the research community to much more quickly home in on promising therapeutics and discard ineffective ones. While these NCATS projects did not use ITAC funding, they could prove just as useful for combating COVID-19 as any of the IRP endeavors that did.
Other NCATS researchers, however, did make use of money provided by ITAC. For instance, the NCATS 3D Tissue Bioprinting Laboratory, led by Marc Ferrer, Ph.D., is developing 3D models of human lung, intestine, and brain tissue to aid studies of how viruses, including the novel coronavirus, infect the cells found in these organs. The models can also be used to test how well antiviral drugs block infection. And because they are complex, 3D structures made up of multiple cell types like actual human tissue, they have a superior ability to offer insights into how COVID-19 affects the body compared to flat, 2D sets of cells that are all the same type.

Scientists in Dr. Ferrer’s lab use 3D printers like this one to create three-dimensional tissue models that more closely resemble tissues in the human body compared to two-dimensional groups of cells.
“We believe that the advantage of these engineered tissue models for therapeutic development is that by capturing the native physiology of tissues and organs, they will be more predictive of the clinical effects of therapies,” Dr. Ferrer explains.
Already, Dr. Emily Lee, a virologist in Dr. Ferrer’s group, and the NCATS team have found that the ability of different SARS-CoV-2 variants to infect these tissue models is similar to the way they infect the same tissues in patients' bodies. In addition, they have observed that changes to the cellular production of immune system molecules called chemokines during SARS-CoV-2 infection vary greatly in different types of tissue, with models that include cells from the lungs’ tiny air sacs, called alveoli, creating high amounts of chemokines. Discoveries like this could assist doctors in figuring out how to help patients with severe cases of COVID-19 who experience immune system overreactions called ‘cytokine storms.’ Dr. Lee and the NCATS team have also assessed the effects of several drugs on the tissue models, finding that the antiviral remdesivir and several other treatments inhibit SARS-CoV-2 infection in lung models while the controversial treatment hydroxychloroquine does not.
Dr. Ferrer’s team at NCATS is collaborating with the laboratory of IRP senior investigator Jonathan Yewdell, Ph.D., a virologist at the National Institute of Allergy and Infectious Diseases (NIAID), and Steven Coon, Ph.D., at the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), who is helping to analyze the extent to which SARS-CoV-2 replicates inside the different types of cells that make up the tissue models, along with using RNA sequencing to examine how SARS-CoV-2 infection influences the activity of genes in the cells. Moving forward, Dr. Ferrer and his collaborators will explore the impact of SARS-CoV-2 on 3D models of human intestinal and brain tissue.
Enhancing Efforts to Prevent the Spread of COVID-19
Prior to the development of COVID-19 vaccines, public health workers relied on other methods to prevent people from catching the illness. These measures remain extremely important for reducing case counts, and several teams of IRP researchers are working on tools that will assist them.
Aside from vaccination, one of the most effective ways to prevent the spread of an infectious disease and reduce hospitalizations is to make sure that infected individuals are diagnosed quickly so they can be isolated and treated as soon as possible. However, early during the course of a viral disease, there may not be enough virus circulating in the body to detect it. This challenge has prompted IRP senior investigator Alexander Wlodawer, Ph.D., to develop a new way to test for the novel coronavirus.

Like the chimera of ancient Greek mythology, which was made up of parts from a lion, a goat, and a snake, Dr. Wlodawer’s chimeric molecule is composed of three different components of the ACE2 receptor, which the novel coronavirus binds to in order to infect cells.
His group’s test uses a ‘chimeric’ molecule that is made by fusing together typically separate parts of the ACE2 receptor, just like the chimera of Greek mythology is made up of three different animals. The SARS-CoV-2 spike protein fits into cells’ ACE2 receptors like a key into a lock, and this interaction allows the virus to infect a cell. Dr. Wlodawer’s team has already designed several different versions of the chimeric molecule and shown that it binds to the virus’ spike protein just like the natural ACE2 receptor. Now, the group is working on a version of the molecule that is attached to a short string of nucleic acids, which will allow it to be used for ultra-sensitive tests that rely on a technology called polymerase chain reaction (PCR). PCR duplicates the genetic material present in a sample, which enables a test to detect even a small amount of the SARS-CoV-2 virus.
Ultimately, Dr. Wlodawer hopes COVID-19 tests that use his chimeric molecule will be able to detect the virus at much lower levels than current tests, allowing doctors and public health workers to more easily identify infected individuals who have not yet developed symptoms before they can spread the illness to others. Moreover, since the chimeric molecule is based on the ACE2 receptor that the virus naturally binds to in order to infect cells, tests that utilize it are unlikely to be fooled by changes to the virus’ spike protein found in different variants.
Other IRP researchers are more concerned with helping public health officials determine where in the U.S. to send resources like COVID tests and implement other interventions. A team led by IRP senior investigator Alison Motsinger-Reif, Ph.D., has developed an online tool called the COVID-19 Pandemic Vulnerability Index (PVI) Dashboard that integrates data on 10 pandemic-related metrics in each county in the U.S., including the county’s current infection and case rates, how much residents move around, the level of testing in the area, and the number of available hospital beds. These numbers are used to create an overall risk profile for each individual county, called a PVI scorecard, which is continuously updated as conditions change.
“My group has longstanding interests in machine learning for making predictions using big data,” Dr. Motsinger-Reif says. “The data that is available related to COVID naturally lent itself to such an effort.”
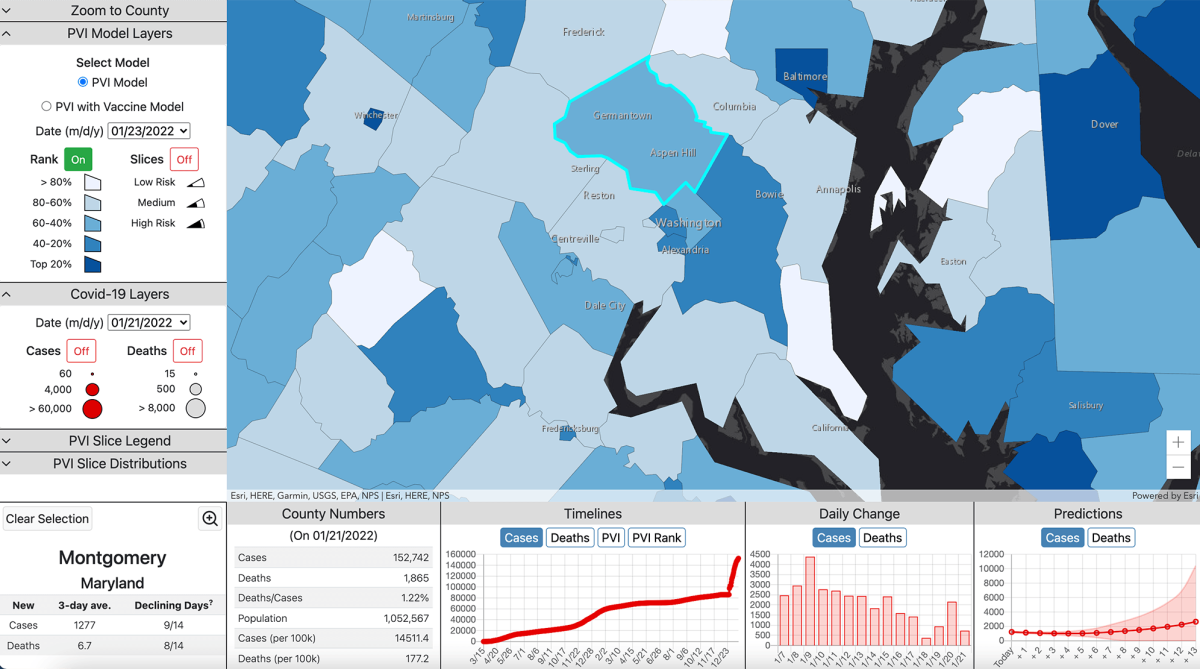
This screenshot from the online tool created by Dr. Motsinger-Reif ‘s lab displays information about Montgomery County, Maryland, where the main campus of NIH is located.
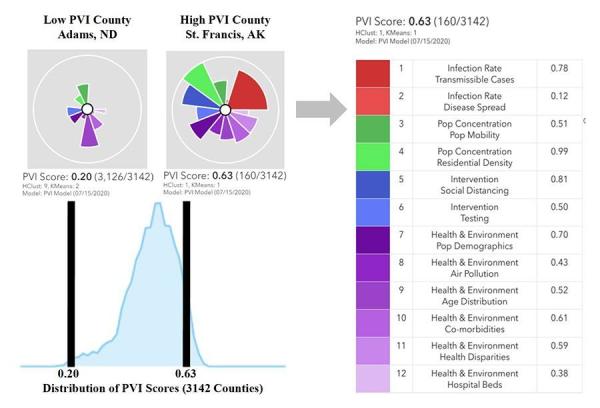
Example of a PVI scorecard for St. Francis County, Arkansas
While numerous similar tools have been created to inform COVID-related decision making, the PVI Dashboard is unique in the large number of datasets it draws from and the clear visual representation it provides of local conditions. Consequently, it is no surprise that it has already helped inform important COVID-related decisions. Public health officials have used it to prioritize distributing masks in certain vulnerable areas of the country, to explain to employees at shuttered manufacturing plants why their plants needed to close while others could remain open, and to support a March 2020 order by the Public Utilities Commission of Ohio that temporarily prohibited gas and electricity supply companies from conducting door-to-door sales. As the team adds even more COVID-related data and statistical modeling tools to improve forecasting of future local case and death rates, the PVI Dashboard will continue to grow even more useful for protecting Americans from COVID-19.
More than two years after the novel coronavirus first appeared, it remains a major threat to human health. However, thanks to new discoveries being made in the IRP and elsewhere, doctors and public health officials are becoming better equipped to face that challenge and save lives.
Subscribe to our weekly newsletter to stay up-to-date on the latest breakthroughs in the NIH Intramural Research Program. And in case you missed it, check out part one of our coverage of ongoing COVID research in the IRP.
Related Blog Posts
This page was last updated on Wednesday, May 24, 2023
