IRP Supercomputer Enables Rapid Response to Coronavirus
Biowulf Lends Massive Computing Power to NIH Research Efforts

Researchers from all corners of the IRP are using NIH’s Biowulf supercomputer to study the virus that causes COVID-19 and identify potential treatments.
Nations around the world are bringing every weapon in their arsenals to the fight against the COVID-19 pandemic: vaccines, new and existing therapeutics, personal protective equipment like face masks, and enough hand sanitizer to fill the Atlantic Ocean. The NIH community is contributing to this unprecedented effort with a tool that no other research institution can claim: Biowulf, the world’s most powerful supercomputer solely dedicated to biomedical research.
Managed and supported by the NIH’s Center for Information Technology (CIT), Biowulf allows IRP researchers and their collaborators to process massive quantities of data with the haste that a global pandemic demands. Between March 23 and August 3, IRP researchers studying COVID-19 used more than 10.2 million CPU hours to process more than 275,000 separate tasks on Biowulf.
“It’s difficult to grasp the significance of those large numbers,” says Steve Bailey, director of high performance computing at CIT. “Suffice it to say that Biowulf is a major scientific instrument, one that enables researchers to understand and make sense of their research data in fields as diverse as genomic analysis, image processing, and statistics.”
CIT has also taken several steps to further increase the speed with which IRP scientists studying COVID-19 can process their data using Biowulf. Prior to the advent of social distancing, the CIT staff held monthly ‘coffee shop consults’ to offer in-person assistance to Biowulf users at convenient locations on the main NIH campus, but these have now been replaced with virtual conferences. With a deep understanding of how Biowulf works, CIT staff continue to advise IRP researchers on how to make effective use of this extraordinary computational resource. In addition, NIH scientists conducting coronavirus-related research are being given ‘priority access’ to the institution’s supercomputing resources. Even a computer as powerful as Biowulf can handle only so many tasks at a time, so when a scientist submits a task to be processed on Biowulf, the request gets put in a queue and a scheduler program determines when to perform it based on a number of factors.
“What we’ve done with COVID-19 priority access is we put those projects at the top of the queue,” Bailey explains. “If someone is doing COVID-19 research, they jump to the front of the line.”
Whether researchers are investigating the structure of the novel coronavirus, scouring databases for potential therapies, or attempting to predict how the disease might spread through communities, the availability of a resource like Biowulf promises to dramatically accelerate the IRP’s contributions to the fight against COVID-19. In fact, the number of coronavirus-related IRP projects that are utilizing Biowulf exceeds the capacity of a single blog post. Read on to learn about a few of these endeavors, and keep your eyes open for another blog in the coming weeks highlighting even more ways Biowulf is helping IRP scientists study this new viral threat.
Searching for COVID-19 Therapeutics
IRP scientists are using Biowulf to simulate how various drugs, natural compounds, and synthetic molecules might interact with proteins on the surface of the novel coronavirus. Image credit: Alissa Eckert and Dan Higgins/CDC
The traditional method of determining whether a compound might be effective at inhibiting or destroying a pathogen is to put the potential drug and its intended target into a petri dish and examine the aftermath. Of course, a single scientist can only run a few of these experiments at a time, making it an extremely slow process. The advent of artificial intelligence and robotics technologies now allow researchers to test much larger numbers of compounds much more quickly via ‘high-throughput screening,’ but even these advances pale in comparison to what scientists can accomplish using Biowulf.
So-called ‘in-silico’ drug screening — a reference to the use of silicon in computer chips — uses a computer to simulate what might happen when two molecules interact. The computer software examines hundreds of random scenarios in which the two compounds come into contact at different places and calculates the energy of the molecular bonds that would form between the compounds in each scenario. If this ‘energy of interaction’ is low enough, the computer considers the interaction as likely to occur in real life, but if that energy is too high, the computer rules it out because the bonds would be unstable or unlikely to form at all. Simulating molecular interactions in this way allows researchers to very quickly evaluate the ability of many different drug candidates to bind to a certain molecule, such as a specific viral protein.
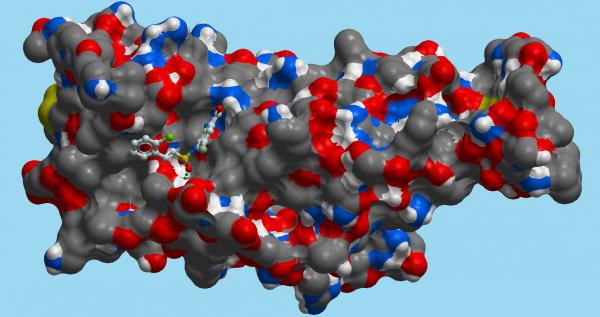
This molecular diagram shows a small molecule docked onto a portion of the novel coronavirus’ spike protein. Supercomputers like Biowulf can accelerate drug development by simulating the stability of interactions between therapeutic compounds and components of the virus.
Researchers led by IRP senior associate scientist Nadya Tarasova, Ph.D., are using Biowulf to sift through thousands of FDA-approved drugs and natural compounds, as well as more than two billion synthetic molecules that can be easily assembled in a lab, in order to determine which ones might be able to bind to various proteins on the surface of the novel coronavirus. Biowulf only requires about 30 seconds to test each compound and can run multiple simulations simultaneously, allowing Dr. Tarasova’s team to evaluate roughly 10 million compounds in a single day.
“We use these virtual screens for drug discovery not only because they are much more economical than actual high-throughput screens, but also because they allow exploration of a much higher diversity of compounds, thus increasing the chances of success,” says Dr. Tarasova.
Her team’s work on Biowulf has already flagged more than 200 molecules as potentially capable of binding to coronavirus proteins, and the researchers have confirmed the supercomputer’s predictions for roughy 100 of them, including several drugs that already have FDA approval for other uses. What’s more, because the simulations being performed on Biowulf examine proteins that are shared by multiple types of coronaviruses, compounds that the IRP team ultimately finds to be effective against the virus that causes COVID-19 could also be effective tools for combating outbreaks of other coronaviruses in the future.
Sussing Out the Structural Details of the Novel Coronavirus
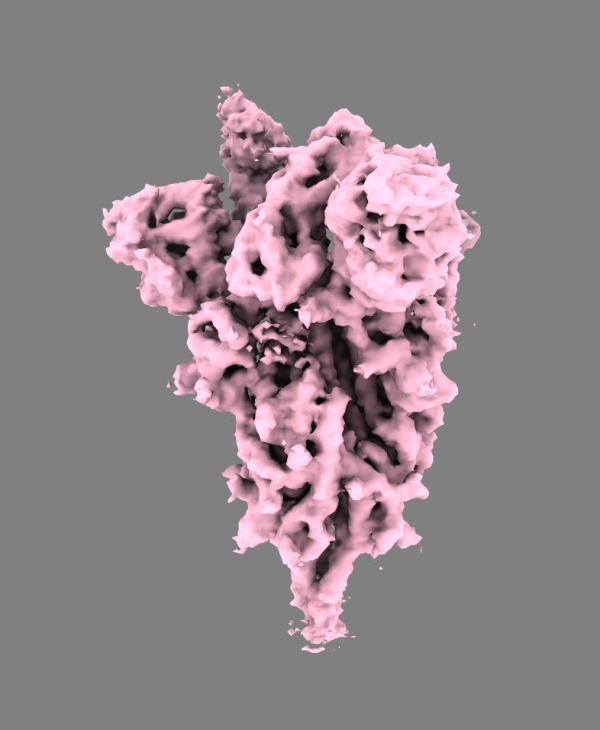
A 3D structural model of the novel coronavirus’ spike protein. Image courtesy of Mario Borgnia
The ability to identify potential therapeutics using computer simulations critically depends on having detailed information about the structure of the novel coronavirus and the proteins that drive its infectiousness. As such, several groups of IRP researchers are using Biowulf to figure out precisely what the virus looks like at the molecular level both by itself and when it interacts with certain proteins.
Researchers in the lab of IRP senior investigator Peter Kwong, Ph.D., for example, are using Biowulf to process images taken using a highly advanced microscope technique called cryo-electron microscopy. They are specifically producing detailed, atomic-level visualizations of the coronavirus’ spike protein, which the virus uses to infect cells. Dr. Kwong’s group is generating images not only of the spike protein itself but also what it looks like when it is attached to the human ACE2 receptor, the gateway by which the virus infiltrates our cells. Meanwhile, a team of researchers led by IRP senior investigator William Copeland, Ph.D., and Mario Borgnia, Ph.D., director of the Molecular Microscopy Consortium at the NIH’s National Institute of Environmental Health Sciences (NIEHS), are specifically focused on examining the outer portion of the coronavirus’ spike protein, known as the ectodomain, since that is the portion of it that initiates contact with cells. Learning more about this specific part of the spike protein could yield useful information about how the virus infects cells and whether lab-produced antibodies can bind to and inhibit it.
Researchers led by senior investigator Markus Hafner, Ph.D., on the other hand, are studying the structural details of the novel coronavirus’ RNA, the genetic material it uses to produce all of the proteins that allow it to function and infect cells. One of the main differences between the RNA genome of the virus that causes COVID-19 and that of other coronaviruses is a short sequence — only 12 base pairs in a genome comprising roughly 30,000 base pairs total — that affects the virus’ spike protein. Amir Manzourolajdad, Ph.D., a research fellow in Dr. Hafner’s lab, theorizes that this minor genetic difference, and others like it, could alter the structure of the virus’ RNA in a way that changes how those genetic blueprints are copied and utilized to manufacture viral proteins. Consequently, developing a detailed structural picture of the novel coronavirus’ RNA could yield insights into the virus’ life cycle and clues as to why it is so infectious.
“Access to a supercomputer like Biowulf makes all these analyses possible,” Dr. Manzourolajdad says. “This is a virus with a long genome, so running these analyses can take anywhere from a few hours up to a week, as well as large amounts of memory. They would be impossible without a supercomputer.”
With a disease that spreads as fast as COVID-19, time saved means lives saved. Consequently, the current pandemic makes clear the value of investments in cutting-edge technology like supercomputing that can dramatically accelerate the pace of research. Thanks to the hard work of hundreds of computing experts at CIT over the past two decades, IRP investigators can take advantage of Biowulf’s impressive computational might to answer key questions about the novel coronavirus more quickly than ever before.
Subscribe to our weekly newsletter to stay up-to-date on the latest breakthroughs about the novel coronavirus and many other diseases from the NIH Intramural Research Program, and keep an eye out for another blog post in the coming weeks exploring still more ways Biowulf is contributing to the fight against COVID-19.
Related Blog Posts
This page was last updated on Tuesday, January 30, 2024
