Research Briefs
Read about scientific advances and discoveries by NIH intramural scientists: detailed atlas for studying Parkinson’s disease neurogenetic risk factors; heart-failure risk significantly higher in rural areas; new mechanism of gene regulation during early development; 3D bioprinting used to create eye tissue; probiotic treatment reduces colonization of harmful bacteria; genotype-first approach uncovers new links to genetic conditions; and first-in-human study evaluates Marburg virus vaccine.
NIA, NINDS: SCIENTISTS HELP CREATE NEUROGENETIC ATLAS FOR STUDYING PARKINSON’S DISEASE
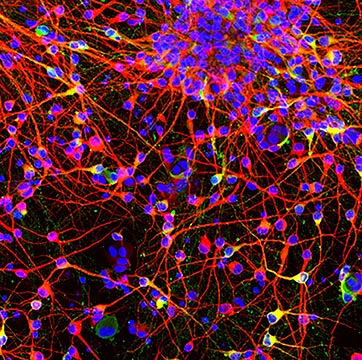
CREDIT: BLAUWENDRAAT LAB, NIA
NIA, NINDS: NIA scientists helped create a detailed atlas for studying Parkinson’s disease neurogenetic risk factors. Advanced genetic data was obtained from cultured dopamine neurons (pictured), the types of cells damaged by the disease.
Scientists at NIA and NINDS took the first steps toward creating a detailed neurogenetic atlas for studying Parkinson’s disease (PD). They made reams of advanced genetic data obtained from 95 cell lines available for researchers to explore online. Creation of the atlas was outlined in an article in Cell Genomics.
“We hope the data provided through this atlas will help scientists worldwide study how Parkinson’s disease genetic risk factors may damage the nervous system and ultimately spur development of new treatments,” said study author and NIA Stadtman Investigator Cornelis Blauwendraat.
The investigators created the atlas as part of the Foundational Data Initiative in PD (FOUNDIN-PD), a joint project among NIH scientists and research centers in Arizona, California, and Germany. The data were obtained from dopamine neurons, the type of brain cells typically damaged by PD. These cells were derived from donated human cells that had been reprogrammed (induced) into pluripotent stem cells and then grown into neurons. Many of the donors were diagnosed with PD or carried a gene variant associated with the disease. In the Cell Genomics article, the scientists described how they grew the neurons and then used advanced molecular analysis techniques to obtain the data. Much of the data provides detailed information about the activity and regulation of PD gene variants.
The cells were donated by volunteers in the Parkinson’s Progression Markers Initiative (PPMI), a clinical study sponsored by The Michael J. Fox Foundation. Scientists can browse atlas data at the FOUNDIN-PD website or download them along with other relevant data, such as brain scans and blood tests, from the PPMI website: http://www.ppmi-info.org. (NIH authors: X. Reed, F.P. Green, A. Bielina, K.J. Billingsley, J. Berghausen, V. Pitz, D. Patel, K. Daida, Y. Li, D.G. Hernandez, M. Nalls, J.R. Gibbs, M.R. Cookson, A.B. Singleton, and C. Blauwendraat, Cell Genomics 3:100261, 2023)
NHLBI, NIMHD: HEART-FAILURE RISK SIGNIFICANTLY HIGHER IN RURAL AREAS
NIH
Adults who live in rural areas of the United States were 19% more likely to develop heart failure (HF) than those living in urban areas, according to a recent study by scientists at NHLBI, NIMHD, and their colleagues at Vanderbilt University Medical Center (Nashville, Tennessee).
Controlling for variables such as smoking and cardiovascular risk factors, the investigators found that the most vulnerable group of rural Americans were Black men, with a 34% higher risk of HF than their urban counterparts. White women had the second highest risk of HF at 22%, which was higher than Black women. For white men, rural living did not correlate with a higher HF risk.
The research team analyzed data from The Southern Community Cohort Study, which enrolled 27,115 adults from 12 states located in the southeastern United States. NHLBI Senior Investigator Véronique Roger, who is also a cardiologist, noted that heart failure can be prevented by following a heart-healthy lifestyle and that one of the biggest contributors to heart failure is high blood pressure, which Black men experience at disproportionately high levels. This study highlights urgency for further investigation, especially the role of structural racism, inequity, and the improvement of interventions that address disease prevention in rural communities. (NIH authors: S.E. Turecamo, T.M. Powell-Wiley, J. Joo, and V.L. Roger, JAMA Cardiol 2023; DOI:10.1001/jamacardio.2022.5211)
NICHD, NCI: SCIENTISTS DISCOVER NEW MECHANISM OF GENE REGULATION DURING EARLY DEVELOPMENT
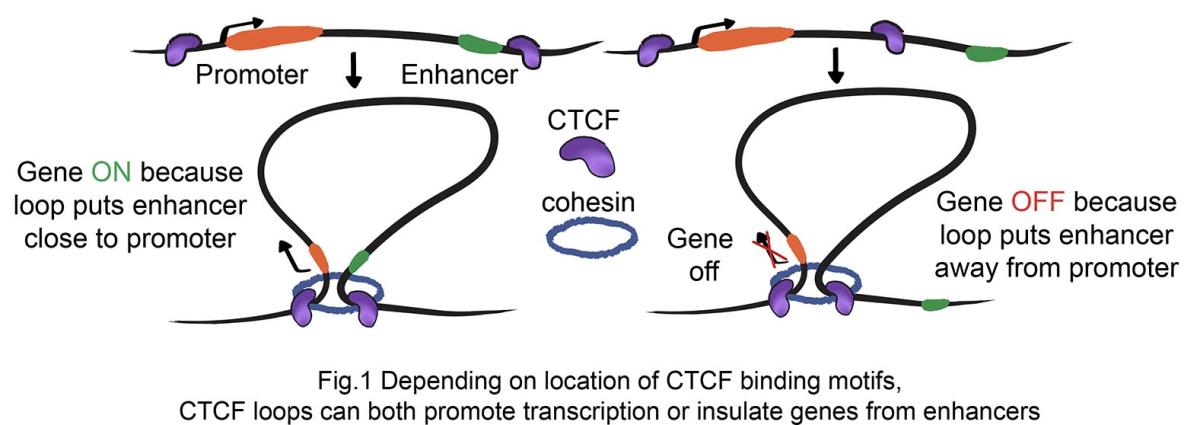
CREDIT: NICHD
NICHD, NCI: Illustration showing how genes can be turned on or off depending on the location of CTCF binding motifs. CTCF loops can both promote transcription or insulate genes from enhancers.
In a growing embryo, the mechanisms by which genes turn on or off to ensure a healthy pregnancy are not completely understood. A recent study by NICHD and NCI investigators revealed that the CTCF protein—which is known to play a role in blocking gene expression—acts strongly in some types of tissues, but not in others. Moreover, the research team also found that the regulation of expression of some genes can bypass CTCF-generated boundaries, providing new insight into the early stages of mammalian development.
One way that cells regulate gene expression is through noncoding strands of DNA known as enhancers that seek out their specific target promotors—a region of DNA where gene transcription can either be initiated or blocked from occurring. Formation of DNA loops by CTCF has traditionally been proposed to block enhancers from interacting with unspecific promoters.
The scientists used mutant mouse models to study how CTCF loops can interfere with expression of the Sox2 gene, which is essential for the accurate embryonic development of many cell types. By inserting CTCF loops between Sox2 and its enhancers, the authors were surprised to find that in some tissues, such as epiblast and neural, CTCF did not repress Sox2 from being activated. However, in other tissues such as the gut, the same CTCF loops completely blocked the enhancers from Sox2, resulting in malformation of the trachea and esophagus. According to the authors, further research is needed to understand why some enhancers are more susceptible than others to regulation by CTCF loops. (NIH authors: S. Chakraborty, N. Kopitchinski, Z. Zuo, A. Eraso, P. Awasthi, R. Chari, A. Mitra, R.K. Dale, T.J. Petros, and P.P. Rocha, Nat Genet 55:280-290, 2023)
NEI, NCATS, NHLBI: RESEARCHERS CREATE EYE TISSUE USING 3D BIOPRINTING
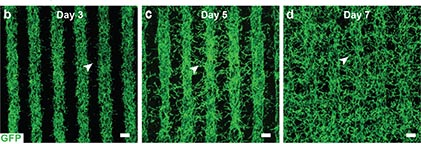
CREDIT: KAPIL BHARTI, NEI
NEI, NCATS, NHLBI: Growth of blood vessels across printed rows of an endothelial-pericyte-fibroblast cell mixture. By day 7, blood vessels fill in the space between the rows, forming a network of capillaries.
Age-related macular degeneration (AMD) is a leading cause of blindness, and understanding the disease’s progression to better treat patients requires accurate and relevant models. Using 3D bioprinting, tissue engineering, and induced pluripotent stem cells, NIH researchers constructed human eye tissue and used it as a new model for studying AMD.
AMD begins in the outer blood-retina barrier (oBRB), which is composed of the retinal pigment epithelium (RPE), Bruch’s membrane, and the choriocapillaris. Those tissue layers provide critical support to the retina’s light-sensing photoreceptors. The new 3D bioprinted eye tissue mimics the structure and function of the oBRB, including a capillary bed.
When the scientists stressed the tissue, they found that it accurately modeled clinical dry or wet AMD, and found that drugs used to treat AMD restored the tissue’s health. In dry AMD, proteins and lipids accumulate beneath the RPE and Bruch’s membrane, whereas in wet AMD, the choriocapillaris hyperproliferates. Both conditions ultimately result in the death of photoreceptors.
The team has now added other cell types to their model, such as immune cells, to even more accurately reconstruct natural eye tissue; and they have built patient-specific tissues using an individual’s own stem cells. This work opens possibilities of stratifying drug trials for specific patient groups and developing patient-specific tissue transplants. (NIH authors: M.J. Song, R. Quinn, E. Nguyen, C. Hampton, R. Sharma, T.S. Park, T. Voss, C. Tristan, C. Weber, A. Singh, R. Dejene, D. Bose, Y. Chen, P. Derr, K. Derr, S. Michael, F. Barone, G. Chen, M. Boehm, A. Maminishkis, I. Singec, M. Ferrer, and K. Bharti, Nat Methods 20:149–161, 2022)
Watch a video of how NIH researchers use bioprinting to create eye tissue: https://www.youtube.com/watch?v=BVICpBpOiB4
NIAID: PROBIOTIC TREATMENT REDUCES COLONIZATION OF HARMFUL BACTERIA
CREDIT: NIAID
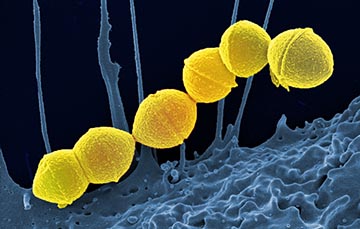
NIAID: Methicillin-resistant S. aureus (yellow) being ingested by neutrophil (purplish blue).
Staphylococcus aureus (S. aureus) is a bacterium that often lives in and on the human body without causing any harm—but it can sometimes cause serious or fatal infections. Some strains such as methicillin-resistant S. aureus (MRSA) are antibiotic resistant, and so other ways to control those pathogens are being explored. A new study shows how the use of probiotics—live microorganisms associated with several health benefits—could be an effective preventative strategy.
NIAID researchers and their colleagues assessed how well the probiotic Bacillus subtilis (B. subtilis) reduced the ability of S. aureus to colonize the human gut and nose. They enrolled 115 healthy adults who were naturally colonized with S. aureus in their intestine and nose and were given a probiotic treatment or a placebo once daily for 30 days. The probiotic led to a 97% reduction of S. aureus in the stool, which reflected intestinal colonization, and a 65% reduction in the nose, compared with no change in the placebo group. Moreover, probiotic administration did not alter the intestinal microbiome, which is a common adverse side effect of using antibiotics and antiseptics.
The findings suggest that B. subtilis could be a promising treatment strategy to diminish S. aureus colonization due to its effectiveness and safety. The authors note that while B. subtilis is a transient colonizer, repeated application to maintain low S. aureus concentrations is not a problem due to the absence of side effects. The research team plans to test the probiotic in trials to see whether the approach can be used to prevent infection. (NIH authors: P. Piewngam and M. Otto, Lancet Microbe 4:E75-E83, 2023)
NHGRI, NIAID: GENOTYPE-FIRST APPROACH UNCOVERS NEW LINKS TO GENETIC CONDITIONS

CREDIT: JULIA FEKECS, NHGRI
NHGRI, NIAID: Doctors researching DNA and genetics.
NHGRI researchers and their collaborators have demonstrated the power of preventative genomic medicine through an approach the scientists call reverse phenotyping. Rather than deducing the genotype from a patient’s clinical findings, the phenotype is determined from the analysis of individuals with specific genomic variants, and then evaluating their physical characteristics.
The research team described three types of discoveries made by analyzing 13 NIH studies that piloted the new approach: In one study the NHGRI investigators in collaboration with NIAID identified new links between the TPSAB1 gene and symptoms affecting the gastrointestinal tract, connective tissue, and nervous system; another study uncovered previously unknown symptoms related to a known genomic variant associated with a metabolic disorder; and a third study found that a genomic variant was associated with immune system dysfunction, potentially helping clinicians understand newly described disorders.
NIH intramural researchers now have access to the data through the Reverse Phenotyping Core Genomic Data Browser database to help identify genomic variants of interest in their own work.
“Studying how taking a genotype-first approach to research can help us learn how to model predictive and precision medicine in the future,” said senior author Leslie Biesecker. In contrast to only treating patients with observable symptoms, this new technology has the potential to help physicians diagnose conditions that are latent and potentially beneath the surface. (NIH authors: C.M. Wilczewski, J. Obasohan, J.E. Paschall, S. Zhang, S. Singh, M. Similuk, T.G. Wolfsberg, C. Turner, L.G. Biesecker, and A.E. Katz, AJHG 110:3-12, 2023)
NIAID: FIRST-IN-HUMAN STUDY EVALUATES MARBURG VIRUS VACCINE
CREDIT: NIAID
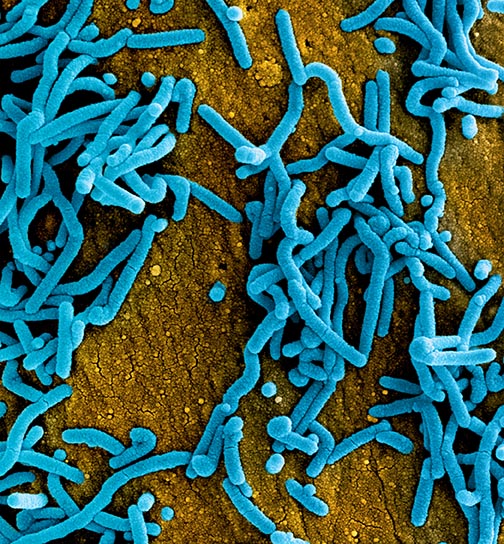
NIAID: Colorized scanning electron micrograph of Marburg virus particles (blue) both budding and attached to the surface of infected VERO E6 cells (orange).
Marburg virus (MARV), like Ebola virus, is a filovirus that causes severe hemorrhagic fever and can be lethal in up to 90% of those infected. And new outbreaks are occurring: Ghana reported its first cases in 2022, and as recently as February 2023, Equatorial Guinea confirmed its first-ever outbreak of MARV. There are currently no FDA-approved vaccines to prevent the disease. In a first-in-human trial, an experimental vaccine developed by NIAID scientists at the Vaccine Research Center was found to be safe, well-tolerated, and effective.
Known as cAd3-Marburg, the vaccine uses an inactivated virus that displays a glycoprotein found on the surface of MARV to elicit an immune response.
The study enrolled 40 healthy adults at the Walter Reed Army Institute of Research Clinical Trials Center (Silver Spring, Maryland) who received either a high or a low vaccine dose. They were then regularly monitored over the course of 48 weeks for adverse reactions and to track their immune responses. In both groups, after four weeks a single dose of the vaccine elicited a robust antibody response in 95% of the participants. That response remained durable in 70% of participants after 48 weeks.
According to the authors, the encouraging results are a crucial step in the development of a vaccine that could someday be used to respond to MARV outbreaks. Further trials are planned to test cAd3-Marburg in Ghana, Kenya, Uganda, and the United States. (NIH authors: K.V. Houser, A.R. Hofstetter, A.M. Ortega-Villa, C. Andrews, G.V. Yamshchikov, S. Hickman, M.R. Gaudinski, I.J. Gordon, L.A. Holman, A.T. Widge, L. Strom, M. Happe, J.H. Cox, S. Vazquez, D.A. Stanley, T. Murray, C.N.M. Dulan, R. Hunegnaw, S.R. Narpala, P.A. Swanson II, M. Basappa, J. Thillainathan, M. Padilla, B. Flach, S. O’Connell, O. Trofymenko, P. Morgan, E.E. Coates, J.G. Gall, A.B. McDermott, R.A. Koup, J.R. Mascola, A. Ploquin, N.J. Sullivan, J.E. Ledgerwood, and the RV507 Study Team, Lancet 401:294-302, 2023)
This page was last updated on Thursday, March 16, 2023
