2018 Research Festival: Selected Concurrent Symposia
The Chemists
NIH chemists wield powerful tools that probe biological processes, decipher the makeup of molecules, and create a foundation upon which treatments for diseases ranging from cancer to HIV/AIDS are derived. Some of the chemists were on hand at the Research Festival to share their success stories.
“There’s such a diversity of chemists [at NIH] even though there aren’t very many of us,” said Amy Newman (NIDA), a mild-mannered chemist who does addiction research using target-directed fluorescent molecules and a fierce-sounding technique called STORM (short for direct stochastic optical reconstruction microscopy). In her search for potential therapies for substance-use disorders, she combines state-of-the-art synthetic organic chemistry techniques with molecular modeling to study dopamine transporters (DAT) and interpret pharmacological data. “We’ve been developing atypical DAT blockers to treat cocaine- and methamphetamine-use disorders,” she said.
Chemists also rely on several techniques to probe the structure and function of the nucleic acids, RNA and DNA. For the past few years, John “Jay” Schneekloth Jr. (NCI) has been using small-molecule microarrays to screen a chemical library of some 20,000 compounds in hopes of finding therapeutically relevant nucleic-acid targets. With the help of nuclear magnetic-resonance spectroscopy and other methods, he has discovered details about the structure of RNA and DNA. Such information is critical for understanding and designing inhibitors, he explained. He recently identified a druglike compound that binds to the HIV transactivation response hairpin and is planning to identify more small molecules that selectively interact with other RNA targets.
CREDIT: LAUREL HOCHSTETLER
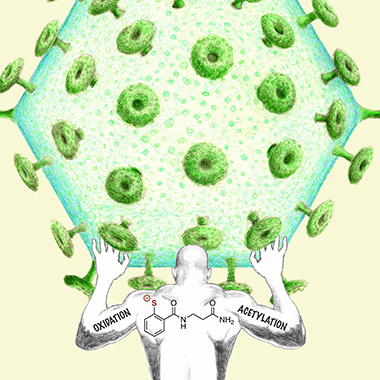
Dan Appella has synthesized two mercaptobenzamide molecules that inhibit the maturation of HIV by a unique mechanism that is highly unlikely to lead to viral resistance. This image depicts the critical role of both oxidative and acetylative mechanistic pathways in one of the molecules (thiolate of SAMT-247) in its attempts to hold an HIV viral particle at bay. This image was featured on the cover of Chemistry in July 2018. (Chemistry 24:9485-9489, 2018;. DOI:10.1002/chem.201801253)
But it’s hard for chemists to keep up with HIV, which has managed to outwit the drugs used to treat it by developing resistance to them. Those drugs, used alone or in combination, target several parts of the viral replication cycle. Dan Appella (NIDDK), however, has synthesized two mercaptobenzamide molecules that are “hitting some other aspect of the HIV replication cycle” and may have found an HIV weak spot. The mercaptobenzamides inactivate HIV’s nucleocapsid protein via an acyl transfer mechanism. So far it looks as if HIV is unable to develop resistance to Appella’s compounds, which appear to be nontoxic. He is examining whether these molecules can be further developed into new drugs to combat HIV infection.
Chemists are at work in the arena of neurodegenerative diseases, too. Jennifer Lee (NHLBI) is harnessing the power of biochemical and biophysical techniques to decipher the molecular mechanisms of amyloid formation such as what’s found in the brains of those with Alzheimer, Parkinson, and Huntington diseases. She uses Raman spectroscopy, a method that illuminates a sample with a laser beam that interacts with molecular vibrations and provides structural fingerprints of molecules such as alpha synuclein. She’s observed conformational heterogeneity in the COOH-terminal region of alpha synuclein, a protein involved in amyloid formation. She hopes to, one day, be able to determine points of intervention in which amyloid assembly is linked to pathology.
Another area that chemists are tackling is metastatic cancer and the perinuceolar compartments (PNCs), subnuclear bodies that are increased in metastatic cell lines such as breast, colorectal, and ovarian cancers. “PNC prevalence is a marker of metastatic potential,” said Juan Jose Marugan (NCATS), who led a team that formulated about 150 derivatives of a compound that could reduce PNC prevalence in metastatic cells. The researchers named the best one metarrestin. It could reduce PNC prevalence and kill metastatic cancer cells—including lung, liver, pancreatic, prostate, and breast cancer—in mice. “To make a long story short, we have seen an effect in numerous tumors,” said Marugan. He went on to explain that metarrestin inactivates the PNC by binding with the protein eEF1A2 and inhibiting the function of RNA polymerase and RNA transcriptase. Plans for human trials are underway and may begin at the NIH Clinical Center in 2019.
FARE awardee Michael Luciano (NCI-CCR), in Martin Schnermann’s lab [https://irp.nih.gov/pi/martin-schnermann], is also applying his chemical expertise to finding ways to defeat cancer. He is helping to develop new small-molecule imaging agents that can be used in fluorescence-guided surgery for cancer. The new agents, which are brighter and more chemically stable than conventional fluorophores, illuminate tumor margins and highlight nerves and other sensitive areas that should be avoided during surgery to remove tumors.
The “Chemical Biology” concurrent symposium took place on Thursday, September 13, 2018, and was chaired by Clifton Barry (NIAID) and Joel P. Schneider (NCI-CCR).
Limerick from Jennifer Lee:
Alpha-synuclein’s a disease-related protein,
And its dynamics to learn we are quite keen.
So we’ve ligated two ends,
To learn its twists and its bends.
And kinetic intermediates can be seen.
Genome Editing
Applications, Challenges, and Innovations with CRISPR
CREDIT: ERNESTO DEL AGUILA III, NHGRI
CRISPR-Cas9 is a customizable tool that lets scientists cut and insert small pieces of DNA at precise areas along a DNA strand. This lets scientists study our genes in a specific, targeted way.
Several researchers presented their work using CRISPR-Cas genome-editing technologies and newer forms of genome editing that may one day lead to advanced therapies for diseases.
Visiting fellow Sergey Shmakov (NCBI), in Eugene Koonin’s group, described how computational approaches to explore the diversity of CRISPR-Cas systems allowed him to identify six new type II CRISPR-Cas subtypes that have the potential for becoming novel genome-editing and regulatory tools.
And immunologist Franziska Petermann (NIAMS), a fellow in John O’Shea’s lab, explained that thanks to CRISPR-Cas9, she determined that the long, noncoding RNA NeST regulates interferon gamma expression as a cis-acting enhancer by inducing chromatin-loop formation. NeST is also known as IFNG antisense RNA 1 and Nettoie Theiler’s Pas Salmonella.
NHLBI’s Transgenic Core Facility, led by Chengyu Liu, is using CRISPR-Cas9 techniques to generate mutant mice used by researchers like Lothar Hennighausen (NIDDK). Liu described how Hennighausen’s lab examined mutations at CRISPR target sites (those sequences that researchers intend to alter) and throughout the entire genome using whole-genome sequencing. Whole-genome sequencing provided evidence that CRISPR-Cas9 does not do any damage such as introducing mutations outside the target site.
But “CRISPR editing frequently wreaks genomic havoc at sites it is supposed to cleanly mutate, thus limiting its usefulness,” said Henninghausen. [For an explanation of the problems, see Nature DOI:10.1038/d41586-018-05736-3 (2018)].
To offset some of the problems with CRISPR-Cas9 editing, researchers are searching for new genome editors. FARE awardee Hye Kyung Lee (in Hennighausen’s lab) shared her work in which she used deaminase base editing, a newer form of CRISPR-Cas9-mediated editing. Lee analyzed the genome of mutant mice generated using cytosine base editors (CBE) and an adenine base editor (ABE) and found that the editors have different fidelities, with ABE being more specific than CBE.
Although CRISPR-Cas9 has its limitations, it is being widely used in many areas of both basic and translational research. Jens Kalchschmidt (NIAMS), a fellow in Rafael Casellas’ lab, described how he used CRISPR-Cas9 to study the mammalian Mediator complex, which regulates transcription by RNA polymerase II. Kalchschmidt used CRISPR-Cas9–guide-RNA-based screening to perform detailed structural and functional analysis of the complex’s 33 subunits.
Kyung-Rok Yu (NHLBI), a postdoctoral fellow in Cynthia Dunbar’s lab, uses CRISPR-Cas9-mediated editing to engineer the nonhuman primate stem and progenitor cells (HSPC) and study aging and advanced therapies for hematologic diseases. He also described his work aimed at making chimeric antigen receptor T-cell (CAR-T) immunotherapy possible for people with acute myeloid leukemia. Cluster of differentiation 33 (CD33) knock-out normal HSPCs generated by CRISPR-Cas9-mediated editing are resistant to CD33-targeted immunotherapy, allowing CAR-T to attack CD33+ leukemia cells without wiping out normal bone-marrow function.
The concurrent symposium “Cell-based Gene Editing and CRISPR” was held on September 12, 2018, and chaired by Lothar Hennighausen (NIDDK).
A Single Cell Can Tell Us a Lot
High-throughput sequencing has transformed research and patient care and paved the way for the personalized-medicine revolution. Now that we can isolate and sequence single cells, we can see what is happening within discrete cell populations in normal or diseased tissue and how this process changes over time.
An example of this approach was presented by Belinda Hauser (NIDCR), a postdoc working with Matthew Hoffman. Hauser is using single-cell RNA sequencing to define cell types during salivary gland development. She hopes this information can be used to help regenerate gland function in patients with head and neck cancer, who have been treated with radiation that often destroys their salivary glands.
FARE awardee Lingling Miao (NIAMS), a postdoc in Isaac Brownell’s lab, is also using single-cell sequencing to define a developmental process in skin. Miao studies Merkel cells, touch sensors that are especially abundant in areas such as the fingertips, lips, and face. By carefully isolating this tiny subpopulation of epidermal cells from mouse skin, she identified discrete subsets of cells at different stages of differentiation, including those that cluster with keratinocytes and likely represent early Merkel-cell precursors.
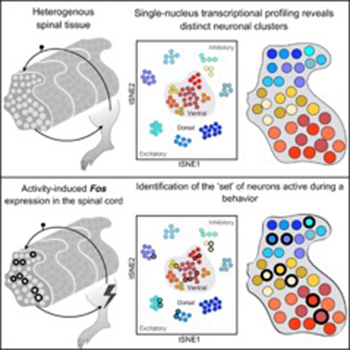
Ariel Levine used single-nucleus RNA sequencing to probe spinal cord cell types in mice. By using this approach after a sensory and a motor behavior, the researchers were able to detect and molecularly identify activated neurons associated with each function. (From A. Sathyamurthy, K.R. Johnson, K.J.E. Matson, C.I. Dobrott, L. Li, A.R. Ryba, T.B. Bergman, M.C. Kelly, M.W. Kelley, and A.J. Levine, Cell Reports 22:2216–2225, 2018; DOI:10.1016/j.celrep.2018.02.003)
Representing an even smaller biological fraction, single-nucleus RNA sequencing is being performed by Ariel Levine (NINDS) and colleagues to map neuronal activity in spinal tissue in mice. Nuclei can be isolated from frozen tissue, which ensures that cellular activity at the time of the experiment is preserved. She hopes what she learns will one day help people who’ve had a stroke or spinal cord injury.
Single-cell RNA sequencing can also be leveraged to develop treatment strategies for individuals with rare syndromes. Keisuke “Chris” Nagao (NIAMS) described a patient with drug-induced hypersensitivity that failed to respond to all conventional treatments. After comparing gene expression between the affected patient and healthy control subjects, Nagao detected in the patient an expansion of certain memory T cells that had a high expression of the JAK3 and STAT1 genes and reactivation of human herpesvirus 6. He found that the patient improved dramatically when tofacitinib, an inhibitor of the protein Janus kinase, and valganciclovir, an antiviral agent, were administered.
Robert Hufnagel (NEI) also discussed how high-throughput exome sequencing can help drive disease-gene discovery in rare neurodevelopmental disorders. He equated his technique to “a magnet trying to pull out the needles from a very large haystack” to narrow the list of potentially causative genetic variants. By using this approach in a cohort of 128 patients with neuro-ocular syndromes, he was able to implicate variants in the UBA2, CSDE1, and SUN1 genes, independently confirmed the association in additional families, and used zebrafish (Danio rerio) CRISPR genetically engineered models to validate his findings.
Another example of how the power of high-throughput technologies can be harnessed to improve patient care was presented by Jun Wei (NCI-CCR). His group is trying to tailor treatments for children and adults with cancer. He described how NCI’s ClinOmics program, which uses genomic approaches to help guide the treatment of NIH Clinical Center patients with cancer, enables precision therapy, provides exome- and deep-targeted cancer-gene-panel sequencing of normal and tumor tissue. These data have been used to provide molecular profiles of 81 diagnoses for 255 NCI-CCR patients.
The “High-Throughput Sequencing–From Single Cell Sequencing to Changing Patient Care” concurrent symposium was chaired by Amy Hsu (NIAID) and Matthew Kelley (NIDCD).
Machine Learning, Data Science, and Hackathons:
The Future Is Now

CREDIT: ALEX LMX, THINKSTOCK.COM
Machine learning, deep learning, and other artificial intelligence technologies are no longer only in the movies. Science fiction is coming to life in projects all over the NIH. In machine learning, computer systems automatically learn from experience without being explicitly programmed. A computer program analyzes data to look for patterns, determines the complex statistical structures that identify specific features, and then finds those same features in new data.
Francisco Pereira, who leads a machine-learning team in the National Institute of Mental Health, described how his team is using various machine-learning techniques and brain imaging (spectroscopy and functional, structural, and diffusion magnetic-resonance imaging) to address research questions in clinical and cognitive neuroscience. For example, individual differences extracted from functional magnetic-resonance-imaging data can be used to predict which drugs may help treat particular conditions. The team also guides researchers in the use of appropriate tools and methods and is developing training activities on basic machine learning.
Noam Auslander described work that she did when was a grad student in Eytan Ruppin’s lab (NCI). She used machine learning to predict patient response to immunotherapy. She developed an immunopredictive scoring system using parameters chosen only from spontaneously regressive neuroblastomas along with their corresponding biomarker measurements. The algorithm successfully predicts patient response to immunotherapy, outperforming current predictive techniques.
Zhiyong Lu (NLM) is helping to make PubMed—NLM’s search engine better. It provides free access to over 28 million biomedical and life-sciences publications. Lu, who researches literature retrieval, natural language processing, and medical imaging through advanced machine learning (deep learning) techniques, has helped improve author name disambiguation (distinguishing authors from others who share the same name) and “most relevant” search results. His machine-learning work has allowed PubMed users to search for relevance-based results by choosing the new sort option called “Best Match.” He also described how PubTator (a web-based tool for automatically annotating PubMed articles with biomedical concepts such as gene and disease names) speeds up manual data curation in NIH-funded GWAS Catalog and UniProt databases by integrating text-mined results. Similar text-mining methods are being used in projects to harvest disease-gene-mutation relationships from electronic medical records, including those containing medical images.
Ke Yan, a postdoctoral fellow in Ronald Summers’ lab the Clinical Center’s Radiology and Imaging Sciences Department, described using deep learning (a type of machine learning based on artificial neural networks and exposure to large data sets) to mine clinical records for computer-aided diagnosis. Well-labeled data is key for deep learning, but annotating medical images requires extensive training, so large data sets for training machine learning are rare. Yan and his colleagues used various strategies to address the problem. For example, disease keywords and phrases were extracted from radiology reports and mapped to their corresponding X-ray images to build a large, fully annotated data set, ChestX-ray14, which contains more than 100,000 chest X-ray images. The dataset enables automated disease classification and localization from X-rays. They further used retrospective clinical annotations to generate the DeepLesion dataset, which has more than 32,000 annotated lesions identified on computed tomography images.
The end of the symposium featured highlights from the NIH Research Festival Hackathon, which brought teams together over three days to tackle computational problems of interest to the intramural research program. Allissa Dillman (NLM) explained how several hackathon teams, each made up of five to six individuals with varying backgrounds and expertise, built machine-learning-based tools to analyze large biomedical datasets including text, images, proteomics, and metadata with a specific emphasis on using machine learning when applicable.
Participating in a hackathon provides scientists with opportunities to network and build programming skills. It also helps people learn best data practices and stewardship, including application of the FAIR principle (that data be findable, accessible, interoperable, and reusable). During the hackathon, one team conducted a live demo of their software and four teams began work on manuscripts. Information about future hackathons can be found at the NCBI Github page.
The “Computational Biology, Data Science, Machine Learning, and Hackathon Results” concurrent symposium was held on Thursday, September 13, 2018, and co-chaired by Matthew Guay (NIBIB) and Eytan Ruppin (NCI-CCR).
CREDIT FOR THUMBNAIL: ROMOLO TAVANI, THINKSTOCK.COM
This page was last updated on Wednesday, April 6, 2022
