RNA-Targeting Therapeutic Restores Protein Absent in Spinal Muscular Atrophy
New Approach Could Enhance Existing Treatments for Debilitating Genetic Disease

A treatment developed by IRP and Oxford University researchers can boost levels of the protein missing in individuals with spinal muscular atrophy by interacting with specific RNA molecules, according to a new study.
The prospect of editing our DNA to treat genetic diseases may have captured the imaginations of scientists and the public in recent years, but that doesn’t mean there aren’t other ways of combating these illnesses. Many promising therapies act not on DNA itself but rather on DNA’s often overlooked cousin, RNA. For instance, experiments in cells performed by IRP researchers have shown promising results for a RNA-targeting therapeutic developed to treat the debilitating genetic disease spinal muscular atrophy.1
The genes distributed throughout our DNA are used as templates to create messenger RNA (mRNA), which cells then use to manufacture the proteins they rely on to survive. In many genetic diseases, a change in a gene leads to the creation of a defective protein or stops the synthesis of the protein entirely; in spinal muscular atrophy (SMA), mutations in both copies of the SMN1 gene cause a lack of the survival motor neuron (SMN) protein. Without the SMN protein, neurons involved in movement die and muscles begin to degrade.
The symptoms of SMA depend in part on how many copies of another gene, SMN2, patients have in their cells. Patients with more copies of SMN2 tend to develop milder symptoms later in life, whereas those with few copies develop severe symptoms, including life-threatening paralysis, at an early age. This is because the SMN2 gene is nearly identical to the SMN1 gene, so it can produce the SMN protein. Unfortunately, small differences between the two genes mean that SMN2 cannot fully compensate when both copies of SMN1 are faulty.
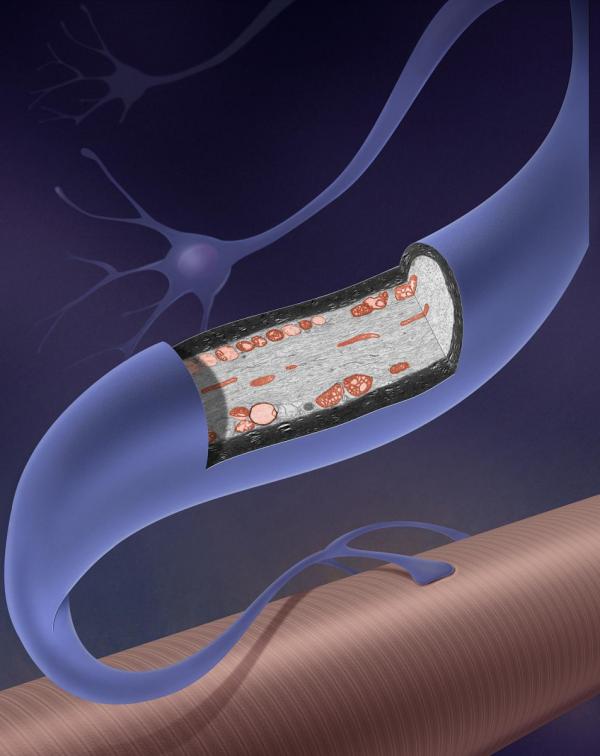
When motor neurons like this one don’t have enough SMN protein, they die and the muscles they connect to begin to atrophy.
“Only a minority of the protein that’s produced by the SMN2 gene is the fully functional form,” explains Audrey Winkelsas, D.Phil., the new study’s first author and a postdoctoral fellow in the lab of IRP senior investigator Kenneth Fischbeck, M.D. “That’s why it’s a good therapeutic target: if we can change how much functional protein is made from SMN2, then that, in theory, could affect disease severity.”
“That’s been shown in animal studies to be very effective at blocking disease onset and mitigating the disease’s manifestations,” adds Dr. Fischbeck, the study’s senior author.
Two of the existing treatments for SMA, nusinersen and risdiplam, change the way SMN2 mRNA is created, resulting in more SMN2 mRNA that can produce fully functional SMN protein. Nusinersen is a type of molecule called an antisense oligonucleotide (ASO), which are strands of RNA-like genetic material that can bind to specific sequences on RNA molecules and cause certain effects depending on their structure. Dr. Winkelsas and Dr. Fischbeck, in collaboration with colleagues at the University of Oxford in the UK, developed several ASOs designed to affect SMN2 mRNA in a different manner from nusinersen by binding to a different part of the mRNA molecule.
More From the IRP
Blog
NIH Scientists Redesign Neurons to Enable Targeted Therapies
When the IRP and Oxford scientists treated cells taken from SMA patients with their experimental ASOs, they found that the ASOs dramatically increased levels of both the functional and non-functional forms of the SMN protein, whereas cells treated with a control ASO incapable of binding to any RNA molecules showed no such change. The treatment also raised levels of two proteins, Gemin6 and Gemin8, which help the SMN protein do its job. Additional experiments revealed that, unlike nusinersen and risdiplam, the experimental ASOs work by stabilizing SMN2 mRNA, allowing more of it to be used to produce the SMN protein before the mRNA breaks down.
“Our approach is complimentary or additive,” says Dr. Fischbeck. “The idea is to enhance the effect of existing treatments so as to reduce the burden of the disease in patients, particularly in older patients who don’t respond as much to available treatments.”
In fact, the researchers tested that idea by treating cells taken from SMA patients simultaneously with one of their experimental ASOs and another ASO that affects SMN2 mRNA in the same way as nusinersen. The combination treatment, they found, increased SMN protein levels more than the nusinersen-like ASO did on its own, suggesting their approach might indeed be synergistic with existing treatments.

Dr. Audrey Winkelsas, the new study’s first author.
Oxford University has applied for a U.S. patent for the new ASO-based treatment, which would allow the institution to license the technology to any pharmaceutical company that might want to continue developing it into a therapy for SMA. Meanwhile, the new study’s results suggest that expanding our knowledge of how ASOs affect mRNA molecules could lead to new treatments for a wide range of genetic diseases.
“On the whole, if we can understand how we can affect the stability of different RNAs, there may be other diseases where increasing the stability of an RNA would be useful,” Dr. Winkelsas says.
“There are more than 6,600 diseases that have a known genetic cause, and many of them result from loss of function of the protein, so any way you can increase amounts of the normal protein by increasing the amount of mRNA could be beneficial,” Dr. Fischbeck adds. “I’d say there are probably hundreds of diseases that might be amenable to this sort of approach.”
Subscribe to our weekly newsletter to stay up-to-date on the latest breakthroughs in the NIH Intramural Research Program.
References:
[1] Targeting the 5' untranslated region of SMN2 as a therapeutic strategy for spinal muscular atrophy. Winkelsas AM, Grunseich C, Harmison GG, Chwalenia K, Rinaldi C, Hammond SM, Johnson K, Bowerman M, Arya S, Talbot K, Wood MJ, Fischbeck KH. Mol Ther Nucleic Acids. 2021 Jan 5;23:731-742. doi: 10.1016/j.omtn.2020.12.027.
Related Blog Posts
- Tiny Molecules Have Big Potential for Treating Eye Diseases
- Global Scientists Come Together at the National Institutes of Health
- Alzheimer’s Patients Show Traces of Cellular Batteries in Blood
- Overturning the Orthodoxy About the Brain’s Stress Chemical
- Rare Genetic Mutation Links Two Neurological Diseases
This page was last updated on Tuesday, January 30, 2024
