Rare Genetic Mutation Links Two Neurological Diseases
Globe-Spanning Collaboration Connected ‘Viking Gene’ to Dementia and ALS

Image credit: Fezcat via Wikimedia Commons
A man with ALS uses a head-mounted laser pointer to communicate with his wife by pointing to letters and words on a communication board. In 2011, an IRP-led study identified a genetic mutation responsible for many cases of ALS and frontotemporal dementia.
June was an important month in the life of baseball great Lou Gehrig. It was the month he was born and the month he was first picked for the Yankees’ starting lineup. Sadly, it was also the month in 1939 when he was diagnosed with the neurological disease that bears his name — Lou Gehrig’s disease, also known as amyotrophic lateral sclerosis (ALS) — and the month he died of that disease two years later. It is appropriate then that ALS Awareness Day is observed on June 21 as a day of hope for those searching for effective treatments and, ultimately, a cure.
IRP senior investigator Bryan J. Traynor, M.D., Ph.D., a neurologist at the National Institute on Aging (NIA), is one of the people leading that search. Best known for his work unraveling the genetic causes of ALS and frontotemporal dementia (FTD), he led an international consortium of researchers that uncovered a mutation on chromosome 9 that is the most common ‘familial’ cause of both ALS and FTD. In fact, this mutation, which disrupts the function of the C90RF72 gene, is responsible for 40 percent of all familial cases of ALS and FTD in European and North American populations, meaning cases in which a family member also has the disease. The discovery, published in 2011, revolutionized the scientific understanding of neurodegenerative diseases and the relationships between them. It also suggested a potential target for future gene therapies.
“The most common gene prior to this finding accounted for about 12 percent of familial cases and here was a gene that explained nearly half of all the familial cases,” Dr. Traynor says. “I think seeing that number was really the moment when it dawned on me just how big of a finding this was.”

American baseball player Lou Gehrig was diagnosed with ALS on his 36th birthday. The disease subsequently became commonly known as Lou Gehrig’s disease.
ALS is a devastating neurodegenerative disease that results from the death of nerve cells in the brain and spinal cord. This causes muscles to shrink and weaken, leading to difficulty moving, swallowing, and, eventually, breathing. It exists in two forms. ‘Sporadic’ ALS is the most common form, affecting about 90 to 95 percent of all patients, and occurs when someone has the bad luck of being born with a random, disease-causing mutation. Familial ALS, which affects the remaining 5 to 10 percent of patients, occurs when a mutation is passed down through generations.
Previous studies had shown that families with a history of ALS also had a history of FTD, which occurs when nerve cells in the frontal and temporal lobes of the brain are lost, causing changes in behavior and personality. In addition, genetic mapping had linked both conditions to the same stretch of DNA on chromosome 9, which was made up of about 7 million base pairs.
“We were just beginning to get an inkling that those two diseases were related to each other,” Dr. Traynor says. “It was pretty rare, but nonetheless, it was proof-of-concept that you could actually have a gene that caused both FTD and ALS.”
In 2009, Dr. Traynor’s NIH team began a genetic study of ALS in Finland, a country with below-average genetic diversity for historical reasons and an unusually high incidence of ALS. After testing samples from about 300 Finnish ALS and FTD patients and 300 people with neither condition, the IRP scientists noticed a common genetic haplotype that exists in the Finnish population. A haplotype is a set of genetic information that is inherited together as a set from a single parent, rather than having a mix of DNA from both parents. It turned out that the vast majority of ALS and FTD patients across Europe and North America had this Finnish haplotype, which included specific variations of three genes: MOBKL2b, IFNK, and C9ORF72.
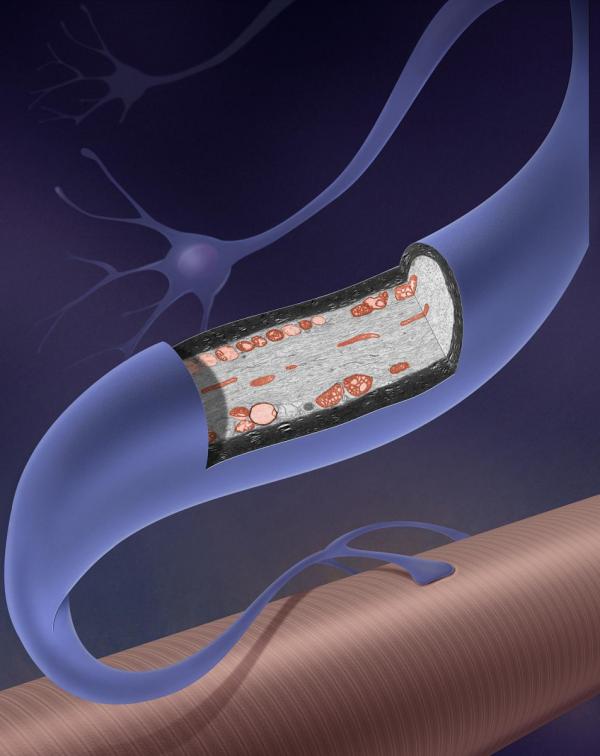
ALS destroys motor neurons like this one that connect to muscle fibers. Without them, movement and vital functions like swallowing and breathing become difficult.
“One of the things about genetics is you can use it as a kind of time machine,” Dr. Traynor says. “What this finding indicates to us is there was probably a one-off genetic event that occurred about 1500 years ago, around the time of the fall of the Roman Empire. We think it was actually the Vikings on their summer holidays who spread this version of the C9ORF72 gene around the rest of Europe.”
Still, even though they had identified the genetic site where a disease-causing mutation must exist and a common set of variations in the genetic profile, researchers had yet to identify the specific gene that was causing all the problems. With a relatively rare disease like ALS, even after narrowing the search to a particular location on a single chromosome, finding a mutation among 7 million base pairs is difficult.
Other research groups all around the world were searching, unsuccessfully, for the mutation, but NIH’s collaborative infrastructure and resources gave Dr. Traynor’s team a competitive edge. They reached out to these other scientists and invited them to send their DNA samples to him for sequencing. He ultimately shared the resulting data for everyone to analyze at the same time, and eventually the globe-spanning team of scientists narrowed the area down to about 200,000 base pairs — “a blink of the eye in genetic terms,” according to Dr. Traynor.
In 2011, Dr. Traynor began searching the DNA samples his team had gathered for variations that didn’t show up in unaffected individuals. This narrowed it down to eight suspects.
“I noticed that six of these eight variations were all very close together — within 30 base pairs — which they shouldn’t have been,” Dr. Traynor recalls. “They should have been distributed randomly across the 200,000 base pairs we were looking at.”
More From the IRP
Blog
Reining in Runaway Enzyme Halts Neuronal Destruction
He compared the samples to a normal ‘reference’ genome and saw that the C9ORF72 gene located in this genetic segment contained multiple repeats of a specific set of base pairs in the patient samples. The reference genome had three repetitions of this genetic sequence within the C9ORF72 gene, but in the DNA from ALS and FTD patients, there were many more. The genetic information was repeated like a bit of song on a skipping record.
“I remember sitting in front of the computer looking at this and that’s when the penny dropped,” Dr. Traynor says. “It was an expansion of six base pairs that were being repeated again and again and again.”
Of course, Dr. Traynor wasn’t the only one hot on the trail of this mutation. The scientific paper announcing his lab’s discovery was published in the journal Neuron alongside a study, led by Mayo Clinic neurogeneticist Rosa Rademakers, Ph.D., that independently linked the same mutation to ALS and FTD, leaving little doubt that the connection was real.
"We’re now beginning to see a grand unified theory of everything coming together from the genomics,” says Dr. Traynor.

IRP senior investigator Bryan Traynor
Since then, Dr. Traynor has continued to make important discoveries about the genetic causes of ALS. For instance, he and his collaborator John Landers at the University of Massachusetts Medical School have identified another gene, called KIF5A, which appears to be involved in damage to the cytoskeleton that provides structure to long neural cells. This type of damage appears to be a significant feature of ALS.
As a practicing neurologist, Dr. Traynor is now focusing his efforts on directing this genetic understanding toward identifying therapies for ALS and FTD. While fixing the C9ORF72, KIF5A, or other mutations through gene editing or CRISPR may be possible one day, Dr. Traynor believes that manipulating the responsible genes’ activity is the most likely direction for therapy right now. He’s excited that such a breakthrough could come in just a few years.
“It's what gets me out of the bed in the morning: just trying to get this done,” Dr. Traynor says. “It's why we're doing what we're doing, why we work at the NIH. We're here to pursue the big ideas.”
Subscribe to our weekly newsletter to stay up-to-date on the latest breakthroughs in the NIH Intramural Research Program.
Related Blog Posts
This page was last updated on Wednesday, May 24, 2023
