Research Briefs
Read about scientific advances and discoveries by NIH intramural scientists: rare gene variants linked to age-related macular degeneration; new approach to treating liver cancer; software assembles complete genome sequences on demand; DNA sequencing during treatment predicts survival from acute myeloid leukemia; a protein crucial to the life cycle of SARS-CoV-2 has a new role; toxic protein that causes a rare muscular dystrophy also kills the precursors of the human nose; autoinflammatory condition reveals therapeutic target.
NEI: ULTRA-RARE GENE VARIANTS LINKED TO AGE-RELATED MACULAR DEGENERATION
CREDIT: NEI
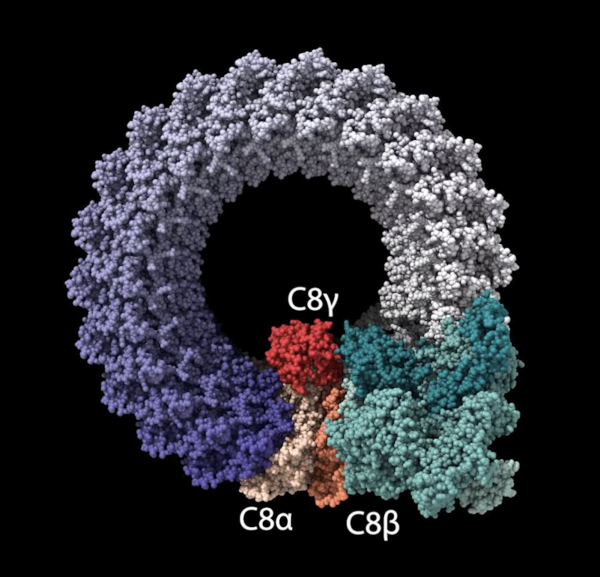
National Eye Institute researchers identified rare genetic variants that could point to one of the general mechanisms driving age-related macular degeneration, a common cause of vision loss in older adults. Shown: Complement factor 8 proteins in the membrane attack complex (MAC). Genetic variants produce malformed C8A and C8B proteins that alter MAC stability, which may drive a chronic inflammatory response in the retina.
Age-related macular degeneration (AMD), a common cause of vision loss in older adults, has several contributing factors, including advanced age, environment, lifestyle, and genetic predisposition. Although there are treatments to slow the progression of some forms of AMD, there is no cure for the condition. An NEI study identified ultra-rare genetic variants that indicate a potential cause of AMD, which could lead to new treatments or preventive therapies.
Researchers examined genetic variants in unrelated families that had multiple members diagnosed with advanced AMD. Individuals with AMD from four such families had mutations in one of two complement factor 8 (C8) proteins: C8-alpha or C8-beta. In addition, investigators reviewed Age-Related Eye Disease Study (AREDS/AREDS2) AMD cohort data, which showed significantly higher prevalence of the C8 variants in that study’s AMD participants compared with the general population.
C8 proteins are part of the terminal stage of the immune system’s complement cascade, a series of inflammatory responses that fight pathogens. The terminal stage of complement pathway forms a ring-like structure called the membrane attack complex (MAC), which helps regulate inflammatory processes in tissues like the retina. The C8-alpha and C8-beta variants affect the ability of the C8 proteins to interact, potentially altering MAC formation and stability. The authors believe that changes in MAC stability contribute to chronic inflammation in the retina, driving AMD progression.
The new findings strengthen understanding of genetic factors linked to AMD pathogenesis and indicate MAC could be a therapeutic target to slow or prevent disease. (NIH authors: L. Zelinger, J. Advani, L. Campello, M.A. English, C. Weber, Y.V. Sergeev, R. Fariss, E.Y. Chew, and A. Swaroop, iScience, 2023; DOI:10.1016/j.isci.2023.106417)
NCATS: SCIENTISTS REVEAL A POTENTIAL NEW APPROACH TO TREATING LIVER CANCER

CREDIT: NCATS
NCATS scientists used the Center’s drug screening capabilities, including drug screening plates like those shown here, to identify a molecule that was effective in killing liver cancer cells. Researchers determined that a specific enzyme was key to turning the molecule into a potential anticancer drug.
Oftentimes in science, the answers you uncover are not the ones you were expecting. Such was the case for NCATS researchers and their colleagues at Massachusetts General Hospital (Boston, Massachusetts) studying cholangiocarcinoma, a cancer arising from liver bile ducts associated with a mutation in the IDH1 gene. After screening thousands of compounds to find a potential therapeutic targeting cells with mutant IDH1, they identified a promising compound called YC-1.
To their surprise, although YC-1 was effective at killing liver cancer cells both in culture and mice models, it had nothing to do with IDH1. Instead, it depended on the sulfotransferase SULT1A1, an enzyme expressed in most liver cells. The investigators discovered that SULT1A1 chemically alters YC-1 through a process known as sulfonation, which converted the molecule into an anticancer drug.
Armed with this new knowledge, the researchers then examined an NCI database of cancer therapeutics to find compounds similar to YC-1 that also required sulfonation to activate their cytotoxic activity. Each compound they found had unique target-binding properties that could target different proteins within a cell—a finding that opens the window into an entirely new class of potential cancer drugs. The scientists published their findings in Nature Cancer after an eight-year collaboration.
The authors note that because different sulfotransferases are expressed in other regions of the body, target compounds could be cherrypicked depending on the type and location of the malignancy, giving doctors yet another precise tool in the war on cancer. (NIH authors: M.I. Davis, K. Kong, T.D. Lee, J.H. Shrimp, W. Zhao, S.E. Kearney, S. Patnaik, M.B. Boxer, M. Shen, and M.D. Hall, Nat Cancer 4:365-381, 2023)
[BY JONATHAN CHU, NIAID]
NHGRI: NIH SOFTWARE ASSEMBLES COMPLETE GENOME SEQUENCES ON DEMAND

CREDIT: ERNESTO DEL AGUILA, NHGRI
National Institutes of Health researchers have developed and released an innovative software tool to assemble truly complete genome sequences from a variety of species.
An innovative new software tool is now able to assemble truly complete genome sequences from a variety of species commonly used in research. Named Verkko, which means “network” in Finnish, the tool was developed by NGHRI scientists and their colleagues, and grew out of assembling the first gapless human genome sequence last year as part of the Telemere-to-Telemere consortium (T2T).
The software compares and assembles different types of genomic puzzle pieces generated by different DNA sequencing technologies, creating an accurate picture of the genome sequence. And although the T2T project took several years to complete, Verkko can finish the task of assembling complete genome sequences in just a few days.
“Now with Verkko, we can essentially push a button and automatically get a complete genome sequence,” said Associate Investigator Sergey Koren, who led the project. The researchers tested Verkko with human and nonhuman genome-sequencing data, and note that the new software will make assembling complete genome sequences as affordable and routine as possible.
Generating gapless genome sequences from various plants, animals, and other organisms will aid in comparative genomics, the study of the differences and similarities among the genomes of diverse species. And with Verkko, scientists can better assess human genomic diversity, such as regions of highly repetitive DNA, across the human population. (NIH authors: M. Rautiainen, S. Nurk, R.P. Walenz, A. Rhie, A.M. Phillippy, and S. Koren, Nat Biotechnol 2023; DOI:10.1038/s41587-023-01662-6)
NHLBI: DNA SEQUENCING DURING TREATMENT PREDICTS SURVIVAL FROM ACUTE MYELOID LEUKEMIA
Acute myeloid leukemia (AML) is a deadly blood cancer and the most common type of acute leukemia in adults. In a recent study, NHLBI scientists demonstrated that testing for the persistence of AML genetic variants after initial treatment predicted survival rates and might be used as a tool to improve long-term outcomes for patients with AML.
The research team used next-generation DNA sequencing to screen the blood of 1,075 adults in remission from AML. All the patients were scheduled to complete treatment by receiving a bone-marrow transplant, which often improves survival outcomes.
At initial diagnosis, 822 of those study participants had mutations in at least one of two genes, FLT3 and NPM1, both commonly associated with AML. After apparently successful initial treatment, the investigators found that 142 of those adults still had residual traces of these mutations despite being classified clinically as in remission. After then receiving a bone-marrow transplant, almost 70% of patients with the lingering mutations relapsed and just 39% survived after three years. In comparison, only 21% of adults without this evidence of trace leukemia relapsed after three years and 63% survived.
The researchers also found that adults with persistent mutations, but who received higher doses of chemotherapy and/or radiotherapy as part of their transplant, were more likely to remain cancer-free after three years than those receiving lower doses. And those adults who didn’t receive stronger treatment as part of their transplant appeared to do better when lower-dose therapy included the chemotherapy drug melphalan. (NIH authors: L.W. Dillon, G. Gui, N. Ravindra, Z.C. Wong, G. Andrew, D. Mukherjee, and C.S. Hourigan, JAMA 329:745-755, 2023)
[BY GUILLERMO RAIMUNDI RODRIGUEZ, NIAID]
NIA: TEAM FINDS NOVEL FUNCTION FOR PROTEIN ESSENTIAL TO SARS-CoV-2 LIFE CYCLE
CREDIT: ROBERT BROSH LAB, NIA. CREATED WITH BIORENDER.COM
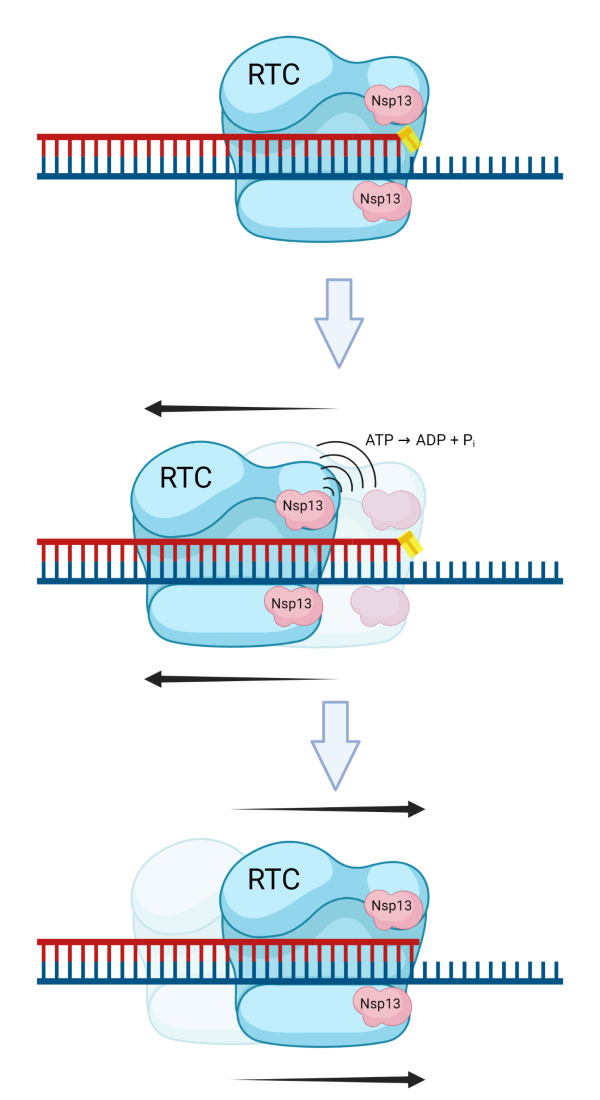
Biochemical studies provide new insight into Nsp13’s role in proofreading of errors that occur during coronavirus replication. Shown: Nsp13 displaces proteins from RNA, a remodeling function not previously attributed to coronavirus helicases.
NIA scientists determined a protein that is crucial to the life cycle of SARS-CoV-2, the virus that causes COVID-19, has a new role. They found that Nsp13 helicase, a protein mainly responsible for unwinding DNA or RNA, displaces proteins from RNA, a remodeling function not previously attributed to coronavirus helicases. Helicases are involved in many biochemical processes in the cell and are found in all living organisms and viruses.
NIA Senior Investigator Robert Brosh led the team that made the discovery using molecular and biochemical techniques. Since age is one of the greatest risk factors for the severity of COVID-19 symptoms, he suggests therapeutics that target Nsp13 could represent an important avenue of translational research that could potentially help older adults recover from COVID-19.
Nsp13’s ability to remove proteins tightly bound to RNA allows the proofreading mechanism of SARS-CoV-2 to correct errors made by the virus’ replication machinery, the system the virus uses to make copies of itself. These mistakes occur when the wrong nucleotides—the basic building blocks of genetic material—are inserted into viral RNA. If left unrepaired, they are incorporated as mutations in future rounds of replication. The authors suggest antiviral drugs that impair Nsp13’s RNA remodeling activity may offer a promising approach to induce a catastrophic level of mutations in SARS-CoV-2, potentially making the virus unable to infect human lung cells. (NIH authors: J.A. Sommers, L.N. Loftus, M.P. Jones III, R.A. Lee, C.E. Haren, A.J. Dumm, and R.M. Brosh Jr, J Biol Chem 299:102980, 2023)
[BY ROBIN ARNETTE, NIA]
NIEHS: TOXIC PROTEIN THAT CAUSES A RARE MUSCULAR DYSTROPHY ALSO KILLS THE PRECURSORS OF THE HUMAN NOSE
NIEHS researchers and their collaborators have found that the transcription factor DUX4 is toxic to the precursor cells of the human nose, and such toxicity may result in arhinia (absence of a nose). Previously, DUX4 has been implicated in a rare type of muscular dystrophy called facioscapulohumeral muscular dystrophy-2 (FSHD2), which causes progressive muscle weakness.
The authors of this study showed that mutations in the SMCHD1 gene resulted in an overproduction of DUX4, killing the precursors of the human nose known as nasal placode cells. The same protein has also been shown to cause muscle cell death in FSHD2.
Using a patient-derived cell line from the two diseases, the authors discovered that expression of DUX4 increased when a cell line was converted to placode cells. Variable degrees of cell death were observed, leading the authors to believe there could be an environmental component that triggered the disease, such as a virus. Indeed, the investigators then demonstrated that cells infected with herpes simplex virus expressed DUX4 in arhinia and FSHD2 cell lines. These findings suggest that the toxic protein may be activated by an environmental modifier, such as a virus, in developing fetuses with genetic risk factors. (NIH authors: K. Inoue, H. Bostan, M.R. Browne, O.F. Bevis, C.D. Bortner, N.P. Martin, S. Chen, A.B. Burkholder, J. Li, and N.D. Shaw, Sci Adv 9:eabq7744, 2023; DOI:10.1126/sciadv.abq7744)
[BY ANNELIESE NORRIS, NCI]
NIAID, NHLBI, NCI, CC, NIAMS: NEWLY DISCOVERED AUTOINFLAMMATORY CONDITION REVEALS THERAPEUTIC TARGET
CREDIT: DAN YANG, NHLBI
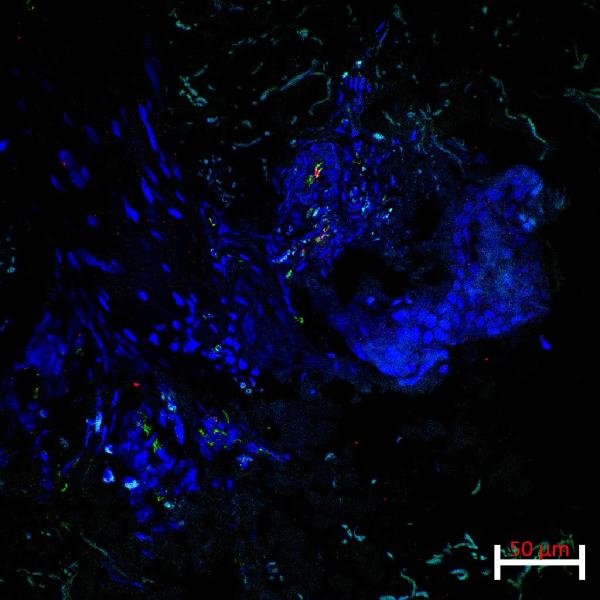
NIH researchers performed experiments to better understand how Lyn kinase interacts with neutrophils and alters inflammatory signals. Shown: Lesional skin biopsy from a patient shows co-expression of the endothelial marker CD31 (green stain) and the activation/adhesion marker, ICAM1 (red stain), in focal areas of a small vessel.
Uncontrolled inflammation as a result of the body attacking its own cells is a hallmark of many autoinflammatory diseases. NIH investigators and their collaborators found that two genetic variants in the LYN gene can cause a newly discovered autoinflammatory disease named Lyn kinase-associated vasculopathy and liver fibrosis (LAVLI).
The scientists identified three pediatric patients: One presented with vasculitis, characterized by inflamed vessels, and the other two also developed liver fibrosis, which is excessive accumulation of scar tissue in the liver. Whole exome sequencing confirmed the mutations in LYN, which encodes for the protein Lyn kinase. Dysregulated Lyn kinase boosts the concentration of neutrophils—white blood cells of the immune system—and can lead to tissue-damaging inflammation.
Next, the researchers performed experiments to better understand how Lyn kinase interacts with neutrophils and alters inflammatory signals. Further drug screening assessments found that TNF inhibitors effectively reduced systemic inflammation, and that the kinase inhibitor dasatinib resolved liver fibrosis. The findings suggest that Lyn kinase may be a potential therapeutic target to treat LAVLI and other types of liver fibrosis driven by inflammation. (NIH authors: A.A. de Jesus, G. Chen, D. Yang, F. Bhuyan, S. Alehashemi, A.T. Rastegar, K. Uss, L. Kardava, B. Marrero, C.R. Lee, D.E. Kleiner, C.M. Hadigan, S.M. Hewitt, S. Pittaluga, C. Carmona-Rivera, K.R. Calvo, N. Shah, D.L. Fink, S.R. Brooks, R.L. Harper, H. Kuehn, M.J. Kaplan, S.D. Rosenzweig, Z. Deng, S.L. Moir, D.B. Kuhns, M. Boehm, and R. Goldbach-Mansky, Nat Commun 14:1502, 2023; DOI: 10.1038/s41467-023-36941-y)
[BY DARWING PADILLA ROLON, NIAID]
This page was last updated on Tuesday, May 9, 2023
