Gisela Storz, Ph.D.: Discovering Big Secrets in Small Genes
Her colleagues laughed at her “crazy idea” when she was a graduate student at the University of California, Berkeley, in the 1980s. Gisela Storz had predicted that a single protein (OxyR) could sense a destructive oxidant, hydrogen peroxide, bind to DNA, and turn on genes that would neutralize the threat. But Storz has gotten the last laugh. Turns out that her hypothesis was correct.
PHOTO CREDIT: BILL BRANSON
Gisela Storz, a distinguished investigator at the National Institute of Child Health and Human Development and a member of the National Academy of Sciences, does research on the identification and characterization of small, noncoding RNAs and of small proteins made up of 50 amino acids or less.
Storz, now a distinguished investigator at the National Institute of Child Health and Human Development and a member of the National Academy of Sciences, continued to study OxyR throughout her training and later as an investigator at NIH. For many years, her lab studied redox-sensitive transcription factors and the bacterial and yeast responses to oxidative stress. Her group discovered that the activity of the Escherichia coli transcription factor OxyR is regulated by a reversible disulfide-bond formation (Science 279:1718–1721, 1998). Her group also serendipitously found one of the first small regulatory RNAs—the OxyS RNA (Cell 90:43–53, 1997). Today, her lab focuses on the identification and characterization of small, noncoding RNAs (sRNAs) and of small proteins (ones with 50 amino acids or fewer).
Storz highlighted her lab’s many contributions to the field of sRNAs and small proteins at this year’s Anita Roberts lecture held in May 2017. During nearly 20 years of studying sRNAs primarily in E. coli, the Storz lab has shown that sRNAs are part of many response pathways in bacterial cells. More recent work in the lab has indicated that small proteins also play an important regulatory function.
The sRNAs and proteins are often not detected by genetic screens or biochemical assays. Storz noted that many of the sRNAs and small proteins that laid the foundation for this field were discovered by serendipity. It has since been recognized that sRNAs regulate bacterial adaptation to various environmental conditions including cell-envelope stress, iron availability, and carbon metabolism. Adaptation to these environments is often essential for bacterial survival and virulence. Regulatory sRNAs can function in primarily two ways: 1) as protein-binding sRNAs that titrate the activity of proteins and 2) as antisense sRNAs, the more common type, that bind to a target messenger RNA (mRNA), thereby affecting transcription, stability, or translation of the mRNA. In her lecture, Storz focused on the latter class of regulatory sRNAs.
As one example, she described how her lab, with the help of then-postdoc Taylor Updegrove (now in the National Cancer Institute, NCI), collaborated with researchers at the University of California at San Francisco to show that an sRNA called MicL was regulating the outer-membrane lipoprotein lipoma-preferred partner, the most abundant protein in E. coli (Genes Dev 28:1620–1634, 2014).
Most of the sRNA-mRNA systems studied to date were discovered at the level of each individual sRNA, but the Storz lab and others have moved to more-global genomic approaches for identifying sRNAs and their mRNA targets. Sahar Melamed, a postdoctoral fellow in the Storz lab, is currently using a deep-sequencing approach termed RIL-seq that he developed at the Hebrew University of Jerusalem (Jerusalem). The technique allowed him to identify four sRNAs that are regulated by the flagellar sigma factor as well as the mRNA targets of these sRNAs. He has shown that the overexpression of one of these sRNAs results in cells with significantly more flagella.
While studying sRNAs, the Storz lab kept stumbling across small proteins and started to characterize their function. For example, a former postdoc in the Storz lab, Lauren Waters, noted a short open-reading frame in the sRNA RybA and later showed that it encoded a small protein. It is currently hypothesized that the small protein regulates the activity of the manganese exporter. Waters has continued to work on this system in her own lab at the University of Wisconsin-Oshkosh (Oshkosh, Wisconsin).
The small protein that has been most extensively characterized by the Storz lab is AcrZ, which co-purifies with a component of a multidrug efflux pump that confers resistance to a wide variety of antibiotics and other compounds in E. coli. Interestingly, cells lacking AcrZ are only sensitive to a subset of these drugs, suggesting that AcrZ affects the specificity of drug export (Proc Natl Acad Sci U S A 109:16696–16701, 2012).
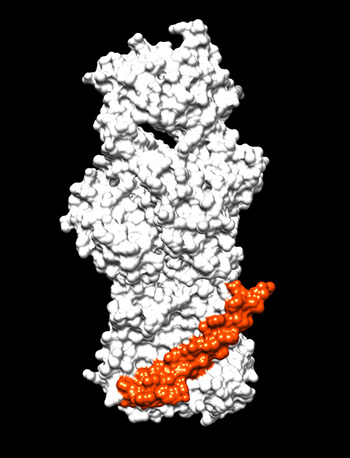
IMAGE CREDIT: HANBO WANG, NICHD
Interactions between small AcrZ protein (orange) and inner membrane component AcrB (white) of a major drug efflux pump in E. coli. Interaction, which takes place in the inner membrane, modulates the specificity of the AcrB-AcrA-TolC transporter.
Storz also explained how she decided to develop more-systematic approaches for finding small proteins. One of her postdocs, Jeremy Weaver, is currently working on a mass-spectrometry approach for identifying novel small proteins in bacterial cells.
At the end of her lecture, Storz took some time to recognize the many contributions of her current and past lab members. In addition, she mentioned some of the pioneering women who were her mentors and who influenced her early in her career at NIH: the late biochemists Thressa Stadtman (National Heart, Lung, and Blood Institute) and Claude Klee (NCI), and her ongoing collaborator, microbiologist Susan Gottesman (NCI). Stadtman was known for her work on anaerobic electron transport and selenium biochemistry. Klee is remembered for her pioneering work on the biochemistry of calcium-dependent signaling and calcium-binding proteins. And Susan Gottesman is well known for her work in the area of regulated proteolysis and the co-discovery and characterization of many bacterial sRNAs.
Their support was subtle, Storz said, but their assistance with some early experiments and invitations to give seminars had a positive impact. Their inspiration was not limited to her science; they also served as role models for achieving balance between career and family life. She noted that these role models all had spouses at the NIH, and the recruitment of two-career couples at the NIH has had a positive impact on many intramural scientists and provides a unique mechanism for improving the retention of women in science.
The “Anita B. Roberts Lecture Series: Distinguished Women Scientists at NIH” honors the research contributions Roberts and other female scientists have made. Roberts, who spent 30 years at NCI before her death in 2006, was known for her groundbreaking work on transforming growth factor–beta. To watch a videocast of Storz’s Anita Roberts lecture, “The Hidden Secrets of Small Genes,” held in NIH’s Lipsett Amphitheater (Building 10) on May 16, 2017, go to https://videocast.nih.gov/launch.asp?23286.
GISELA STORZ, PH.D.
NIH Distinguished Investigator, Section on Environmental Gene Regulation, and Deputy Chief of the Cell Biology and Metabolism Program, Eunice Kennedy Shriver National Institute of Child Health and Human Development
Born: Logan, Utah
Grew up: Primarily in Fort Collins, Colorado
Research Interests: The identification and characterization of small noncoding RNAs and small proteins of 50 amino acids or less
Education: University of Colorado, Boulder (B.A. in biochemistry); University of California, Berkeley (Ph.D. in biochemistry)
Training: Graduate and postdoctoral training in the laboratory of Bruce Ames (former NIH scientist), University of California, Berkeley; postdoctoral training in NCI’s Laboratory of Molecular Biology with Sankar Adhya; postdoctoral training in genetics, Harvard Medical School (Boston)
Came to NIH: In 1989 for training in NCI; returned in 1991 as a tenure-track investigator in NICHD’s Cell Biology and Metabolism Branch; appointed tenured investigator in 1999, deputy director in 2008, associate scientific director in 2015, and a distinguished investigator in 2016
What inspires her: A desire for an interesting life full of challenges and thinking; people in science that she admires; and friends that she has made in the scientific community
Outside Interests: Her three kids; exercising; gardening
Little-known fact: Was manager of her son’s travel soccer team for a decade; most of the players have gone on to play soccer in college
Website: https://irp.nih.gov/pi/gisela-storz
This page was last updated on Friday, April 8, 2022
