2017 Research Festival: Selected Concurrent Symposia
Cell-Based Therapies to the Rescue
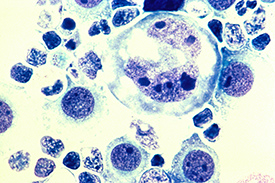
CREDIT: LANCE LIOTTA LABORATORY, NCI (MARCH 1988)
In cancer immunotherapy, T cells can be genetically engineered to treat lymphoma and other blood cancers. Shown: Human lymphoma tumor cells in the pleural fluid magnified to 400x.
Cell-based therapies have been used in clinical trials for more than 25 years with limited effectiveness…until now. Several intramural scientists shared their stories about how cell therapies are contributing to advances in cancer immunotherapy, gene therapy, and regenerative medicine and how the Clinical Center’s Cell Processing Section is playing a critical role.
In cancer immunotherapy, T cells can be genetically engineered to treat lymphoma and other blood cancers. James Kochenderfer’s (NCI-CCR) presentation explained how he developed an anti–cluster of differentiation 19 chimeric antigen receptor (CAR)–engineered T-cell therapy that causes lasting remission in advanced lymphoma. He also created the first anti-B-cell maturation antigen (BCMA) CAR T-cell therapy that can be used to treat multiple myeloma. In addition, his lab is studying how to mitigate cytokine-mediated toxicity and target new antigens.
Also in the gene-therapy realm, Suk See De Ravin (NIAID) described how she and Harry Malech (NIAID) used CRISPR/Cas9 gene editing to correct a genetic mutation in hematopoietic stem cells that causes a rare inherited immunodeficiency disease called chronic granulomatous disease. This targeted gene therapy has shown long-term correction in mice transplant studies. Preclinical studies are underway to determine future clinical applications and whether the therapy could be used to treat other diseases.
Cell-based therapies are also being used to regenerate parts of the eye that are damaged in age-related macular degeneration (AMD). Kapil Bharti (NEI), who has developed an autologous induced pluripotent stem cell (iPS) technology–based cell therapy to treat AMD, described preclinical studies for which he developed an iPS-cell-derived patch that protects against AMD-related vision loss. The patch consists of retinal pigment epithelial (RPE) cells differentiated from iPS cells. Bharti’s team replicated AMD-like RPE damage in pigs and confirmed that the patch could rescue photoreceptor cells from degeneration. The team is currently manufacturing the patch and hopes that by 2018, the patch can be transplanted into people with AMD.
FARE Award winner Defne Bayik (NCI-CCR) is doing research that may have implications for the treatment of autoimmune and inflammatory diseases. She found that targeting certain toll-like receptors (which can recognize pathogens) with an agonist called PAM3CSK4 induces human monocytes to differentiate into immunosuppressive macrophages, thereby reducing the inflammatory response.
It’s the NIH Clinical Center’s Cell Processing Section that produces the cellular therapies used in clinical trials. Section Director David Stroncek (CC) talked about the complexities of producing, analyzing, and developing high-quality cellular therapies. One of the products the section manufactures is CAR T cells, recently FDA-approved for the treatment of children with acute lymphoblastic leukemia. Until recently, CAR T-cell therapy was used only in small clinical trials in people with advanced blood cancers.
The “Cell-Based Therapies: Transitioning from the Research Laboratory to the Clinic” concurrent symposium was chaired by David F. Stroncek (CC).
Searching for What Causes Diseases

The genomics era has led to exciting discoveries in human genetics that have greatly improved the understanding of the causes of rare and common diseases.
Kenneth Fischbeck (NINDS), whose group has identified genes and helped in the development of treatments for hereditary neuromuscular diseases such as Duchenne muscular dystrophy (DMD) and spinal muscular atrophy (SMA), described how oligonucleotides have been used in mice and patients to correct for the disease-causing genetic mutations. Oligonucleotides have a phosphate backbone and have been modified for enhanced stability and activity, Fischbeck said. Two examples are eteplirsen, which was approved last year for some forms of DMD, and nusinersen, a new, effective treatment for SMA.
Douglas Stewart (NCI), who is investigating the genetics and clinical aspects of the rare pediatric lung cancer pleuropulmonary blastoma (PPB), explained how PPB was caused by germ-line mutations in DICER1. DICER1 syndrome increases the risk of a variety of cancerous and noncancerous (benign) tumors, commonly pleuropulmonary blastoma in children but also ovarian and testicular stromal-cell tumors and other rare, cystic neoplasms. Some mutation carriers develop cancer while others remain healthy. Like an astronomer searching for supernova, Stewart said he searches for reasons for clinical variability in the DICER1 syndrome.
In her talk on uncovering etiology and improving patient care through characterization of cancer-predisposition syndromes, Sharon Savage (NCI-DCEG) spoke about her work on the phenotype of dyskeratosis congenita (DC), a disorder that can increase the risk of developing leukemia and other cancers. DC’s characteristic features include abnormal skin pigmentation, oral leukoplakia, and nail dystrophy along with very high risks of bone-marrow failure and cancer. Savage explained that the complicated genetic etiology involves germ-line mutations in telomere-biology genes with autosomal-dominant, autosomal-recessive, and X-linked inheritance.
Daniel Kastner (NHGRI) talked about the expanding spectrum of autoinflammatory diseases and the role of next-generation sequencing in discovering new human diseases. As an example, he discussed how exome sequencing in two unrelated children with unexplained recurrent fevers and early-onset stroke led to his lab’s discovery of mutations in a previously poorly characterized gene called CECR1, which causes the deficiency of adenosine deaminase type 2 (DADA2). With the benefit of genetic testing, many more patients have been recognized with this condition, which can present not only with recurrent stroke, but also with vasculitis or bone-marrow failure. Kastner also discussed his team’s favorable experience with tumor-necrosis-factor inhibitors in preventing strokes in DADA2 patients.
Genomics is showing that the phenotypes associated with neurological diseases are much broader than we previously appreciated. “Genetics is already leading to a reclassification of neurological diseases,” said Bryan Traynor (NIA). “In the longer term, machine learning applied to genomic data will further enhance our ability to recognize and diagnose diseases and to identify individuals within the population at greater risk of disease so that they can benefit from early intervention.”
FARE Award winner Michael Leney-Greene (NIAID), a graduate student in Helen Su’s and Michael Lenardo’s lab, described research on patients suffering from a novel disease of autoimmunity caused by mutations in a gene that is a member of the mammalian target of rapamycin complex 1 (mTORC1) regulatory complex that controls protein synthesis. “Given that its expression is restricted to immune cells and primarily T cells, we hypothesize that it is a putative T-cell-specific regulator of mTORC1 activity,” said Greene. “We were able to use this information to recommend sirolimus (an mTOR inhibitor) as a therapy for one of our patients, who saw drastic improvement of their autoimmune symptoms.”
The “Genotyping and Phenotyping” concurrent symposium was chaired by Kenneth Fischbeck (NINDS) and Sharon A. Savage (NCI-DCEG).
Neuroscience and Compulsive Disorders

Whether it be compulsive eating, excessive alcohol consumption, or substance abuse, such behaviors are driven by shared neurocircuitry that we struggle to understand. The NIH Center on Compulsive Behaviors (CCB), a collaboration of researchers from seven institutes, is advancing scientific discovery in the field of compulsive disorders. Several CCB members presented their research at the 2017 Research Festival.
Michael Krashes (NIDDK), who is trying to decipher the neural wiring that underlies hunger, described his work on hunger-driven motivation in rodents. His group investigated agouti-related peptide neurons in the hypothalamus. The neurons, when stimulated, trigger increased appetite and feeding behavior. His group found that when food is accessible, hunger can suppress other motivators such as thirst, anxiety, and social interaction.
To understand the neural circuitry underlying alcoholism, Veronica Alvarez (NIAAA) has focused on dopamine D2 receptors (D2R) in the striatum area of the brain. Usually, alcohol has a sedative effect. But rodents that are vulnerable to high alcohol use have a heightened sensitivity to alcohol’s stimulative effects. It turns out that the heavy-drinker rodents have a lower availability of striatal D2Rs: DRD2-knockout mice experienced enhanced motor response to alcohol.
Yavin Shaham (NIDA) wants to understand the cellular and neuroanatomical mechanisms that underlie drug relapse in addicts who’ve been abstinent. His group used rat models of methamphetamine and relapse after choice-based voluntary abstinence (a rat model of relapse after cessation of contingency management in humans addicted to drugs). His lab members and collaborators found that neuronal ensembles in the dorsomedial striatum and the glutamatergic projection from the anterior insular cortex to central nucleus of the amygdala play an important role in relapse to methamphetamine seeking after voluntary abstinence.
Philip Shaw (NHGRI) explores the similarities and differences in the neural bases of impulsive and compulsive behavioral disorders. He highlighted some notable differences between impulsive disorders (such as attention deficit hyperactivity disorder) that exhibit abnormalities in the brain’s ventral striatum, and a major compulsive disorder (obsessive-compulsive disorder) that shows abnormalities in the dorsal striatum. However, both these disorders shared deficits in the ability to inhibit responses. This deficit is traced to hypoactivity in the brain’s right inferior frontal gyrus.
FARE Award winner Andrew Kesner (NIDA), a graduate student in Satoshi Ikemoto’s lab, explained how an understudied region in the posterior hypothalamus—the supramammillary nucleus (SuM)—may play an important role in motivational processes. Using optogenetics—a technique that involves the use of light to control neurons that have been genetically modified to express light-sensitive ion channels—Kesner showed that mice will press a lever to activate specific neurons in the SuM. In addition, he used microelectrodes implanted in the SuM to record neuron activity and found that the neurons fired when the mouse was seeking a delicious sucrose solution; they stopped firing when the mouse finally received the reward. The finding suggests that neurons in the SuM could play a role in driving reward-seeking behavior.
The concurrent symposium “Neuroscience and Compulsive Disorders” was co-chaired by Veronica Alvarez (NIAAA) and Philip Shaw (NHGRI).
Single-Cell Analysis: An Old Idea with New Tricks

CREDIT: RHODA BAER
Single-cell analysis means figuring out how every cell is different and then using the unique characteristics of each to solve bigger problems. Until now this technology was difficult to manage, but the marriage of biology, computer science, and bioinformatics has transformed this field. At this session, several intramural investigators showcased the latest in single-cell research at NIH.
John Tsang (NIAID), who studies the immune system, described how individual cells (and cell populations) monitor and adapt to environmental signals. He is particularly interested in how the human body’s environment and age can modify the heterogeneity of macrophages, a type of immune cell. Older people have more homogenous cell populations than younger people do; the homogenous cell populations may underlie changes in the immune function as people age.
Detecting disease before symptoms appear is another field with potentially big payoffs. Jamie Diemer (NHGRI), a biologist in Paul Liu’s lab, presented her project that relies on single-cell analysis to detect abnormal myeloid progenitors as a biomarker in transgenic mice before they develop leukemia. Understanding which cells underlie the development of leukemia will pave the way to better detect or stop the leukemia progression before the onset of disease.
In a similar vein, Eli Boritz from NIAID uses single-cell analysis of infected T cells in his human immunodeficiency virus (HIV) research. His lab is on a quest to identify HIV patients’ viral reservoirs (infected cells not actively producing the virus). Often, patients are on antiretroviral therapy (ART) to reduce viral loads (the amount of HIV in the blood), but infected cells are still circulating in the body. If ART is stopped, patients get sick. If the reservoirs had a marker, clinicians could wipe out that small, but specific, cell population in an effort to cure HIV.
Other researchers’ work had a basic-science spin. FARE winner Yihan Wan (NCI), a postdoc in Daniel Larson’s lab, presented her research visualizing active transcription of genes in single cells in real time. “In the past, much has been made of the process of transcription, but we think the off-time tells us more about the regulation of the gene,” said Wan as she displayed a video showing the active locus lighting up and turning off as a gene was transcribed before the audience’s eyes. Michael Kelly (NIDCD), a research fellow in Matthew Kelley’s lab, described the lab’s use of high-throughput single-cell transcriptional-profiling methods to map how hair cells in the cochlea differentiate through developmental time.
“[A]s single-cell datasets are generated and integrated from lots of different tissues, we can get a whole-organism type of atlas,” said Kelly. Such an atlas could be useful for pharmaceutical targeting and biomarker screenings as a new way to detect and treat diseases.
The concurrent symposium “A Diversity of Biological Insights from Single-Cell Analysis across Multiple Diseases” was chaired by Mark Cookson (NIA) and Yong-Chen William Lu (NCI-CCR).
The Microbiome: A Key Primer of the Immune System
CREDIT: DARRYL LEJA, NHGRI
The microbiome is comprised of microorganisms that live in and on us and contribute to human health and disease.
The importance of the microbiome in human health and in disease progression has revolutionized how we think about disease. In the “Microbiome” concurrent symposium, several intramural researchers discussed the importance of the normal flora in priming immunity and contributing to disease pathologies.
The session chair Yasmine Belkaid (NIAID) discussed the interplay between the skin microbiota and the immune system. She described how postdoc Jonathan L. Linehan (NIGMS) observed that the commensal bacterium Staphylococcus epidermidis induced commensal-specific CD8+ T cells in the skin without causing inflammation or tissue damage. Using a nonhuman primate model, Belkaid and Linehan showed that this increased number of CD8+ cells protected against subsequent infections from pathogenic bacteria and promoted tissue repair.
The role of the microbiome in boosting cancer immunotherapy was the focus of visiting postdoctoral fellow Marie A. Vetizou’s research (NCI-CCR) in the laboratory of Giorgio Trinchieri. She is trying to find a way to improve the efficacy of the monoclonal antibody ipilimumab, used to treat melanoma, without increasing its toxicity. Vetizou used a mouse model to show that the drug’s efficacy was lost in a germ-free environment, but that efficacy could be restored by Bacteroides thetaiotaomicron or B. fragilis, bacteria normally found in the human gut. The two species of bacteria also provided protection against colitis associated with anti-cancer treatment.
Julie A. Segre (NHGRI) underscored the advantages of using shotgun metagenomics, a powerful sequencing approach, for the discovery of eukaryotic viruses, which are currently underrepresented in genome databases. She highlighted a recent study that looked at the skin microbiome from healthy volunteers and from people who had a rare immune disorder called immunodeficiency due to dedicator of cytokinesis 8 protein deficiency (DOCK8 deficiency), which causes persistent skin infections and skin inflammations such as eczema. Strikingly, she found that sequences from eukaryotic viruses made up less than four percent of the microbiome of healthy volunteers, but 90 percent of the microbiome of the DOCK8-deficient patients. The viruses found in the patients were tremendously diverse and included novel variants, types, and species.
The role of the gastrointestinal (GI) tract microbiome and microbial translocation in the progression of simian immunodeficiency virus (SIV) infections in nonhuman primates is the focus of Jason Brenchley’s (NIAID) research. SIV is similar to the human immunodeficiency virus (HIV), and SIV-infected Asian macaques (pigtail macaques and rhesus macaques) progress to simian AIDS. Several groups have shown that microbial dysbiosis (an imbalance in the microbiome) in the GI tract is associated with HIV infection in humans, but the cause is unclear. The dysbiosis does not occur in SIV, however. In recent studies, Brenchley has experimentally induced dysbiosis, with targeted antibiotic use, in Asian macaques to determine how the natural course of SIV would be affected. Surprisingly, experimental induction of microbial dysbiosis did not accelerate the natural course of SIV infection. Brenchley is currently exploring whether GI microbial dysbiosis increases the chances of developing an HIV or SIV infection.
Our growing understanding of the interplay between the microbiome and immunity will allow us to better evaluate the role of the microbiome in disease treatment and provide a foundation for future therapeutics that look at altering the microbiome for disease prevention or management.
The “Microbiome” session was chaired by Yasmine Belkaid (NIAID).
This page was last updated on Friday, April 8, 2022
