An Amazing Two-Day Banquet of COVID-19 Research Presentations
The two-day “NIH/FDA COVID-19 Research Workshop,” held virtually in October 2020, showcased what scientists at NIH and FDA are doing to fight SARS-CoV-2, the virus that causes COVID-19. The workshop took the place of the NIH’s annual research festival, which was cancelled this year due to the COVID-19 pandemic.
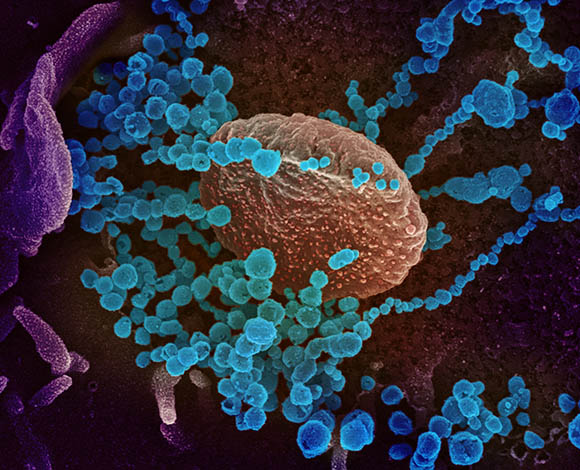
NIAID-RML
This scanning electron microscope image shows SARS-CoV-2 (round blue objects) emerging from the surface of cells cultured in the lab. SARS-CoV-2, also known as 2019-nCoV, is the virus that causes COVID-19. The virus shown was isolated from a patient in the U.S.
Introduction: BY MEGAN KALOMIRIS, NIAID
Session 1: Tracking, Diagnostics, and Prevention, BY FRANCES FERNANDO, NICHD
Session 2: Virology 1: Replication, Evolution, and Host Factors, BY NATALIE HAGEN, NCATS
Session 3: Virology II: Structure, Function, and Inhibition of Viral Proteins, BY SUNITA CHOPRA, NCI
Session 4: Clinical Manifestations and Pathogenesis, BY ETHAN SMITH, NINR
Session 5: Host Response and Immunology, BY JUNE GUHA, NIAID
Session 6: Animal Models and Vaccines, BY CHARLESICE HAWKINS, OITE
Introduction: Sharing COVID-19 Research
October 31st is a date often associated with spooky costumes and eating candy, but it holds additional significance this year—it was the 80th anniversary of President Franklin Delano Roosevelt’s 1940 dedication of the National Institute of Health’s (yes, it was singular back then) Bethesda campus. Roosevelt’s vision for the NIH was to use science to protect the health of the American people, and 80 years later during the midst of the COVID-19 pandemic, the NIH is still doing just that.
To show how much the NIH is using science to fight SARS-CoV-2, the virus that causes COVID-19, the COVID-19 Scientific Interest Group (SIG) organized a two-day workshop where NIH and FDA scientists could share the results of their research. The “NIH/FDA COVID-19 Research Workshop” was held virtually (via WebEx) on October 29 and 30 and took the place of the NIH’s annual research festival, which was cancelled this year due to the COVID-19 pandemic. The SIG worked with National Cancer Institute (NCI) IT operations staff and others to hold a virtual event that attracted more than 1,000 participants who were eager to learn how scientists are tackling this threat to public health.
The event featured roughly 135 presentations including 56 talks and 79 flash talks representing some 250 labs working on COVID-19 projects. National Institute of Allergy and Infectious Diseases Director Anthony Fauci kicked off the first day with an overview of our current knowledge of SARS-CoV-2 and described therapeutic treatments being tested and vaccine trials taking place. In a similar fashion, NIH Director Francis Collins opened the second day by congratulating the researchers presenting their work. The main presentations were divided into six categories: tracking, diagnostics, and prevention; replication, evolution, and host factors; structure, function, and inhibition of viral proteins; clinical manifestations and pathogenesis; host response and immunology; and animal models and vaccines. In addition, flash talks were held for an hour each day, with four sessions occurring simultaneously. These blink-and-you’ll-miss-it presentations were each three minutes long followed by a question or two. Like the main presentations, each flash talk session had its own theme: public health and health disparities; diagnostics; therapeutics; pathogenesis and immunology; data science; virology; and vaccines.

CREDIT: NIAID
Several of the researchers giving presentations work in this Integrated Research Facility at NIAID’s Rocky Mountain Laboratories campus in Hamilton, Montana.
“This was an amazing two-day banquet,” said Deputy Director for Intramural Research Michael Gottesman in his closing remarks at the end of the workshop. He noted that almost all the talks described research that involved collaborations and featured “amazing research talent at NIH.”
With active engagement in the audience and virtually no technical hiccups, the organizers achieved their goal of bringing scientists together.
During his remarks, Collins reflected on the vision President Roosevelt’s had for the NIH 80 years ago. His “dream of how this institution could contribute…against the next worldwide pandemic,” has come true, Collins said. “And you all are part of that.”
Videocasts are available at:
- Day 1, General Session: https://videocast.nih.gov/watch=40137
- Day 1 Flash Talks: https://videocast.nih.gov/watch=40167
- Day 2, General Session: https://videocast.nih.gov/watch=40138
- Day 2 Flash Talks: https://videocast.nih.gov/watch=40168

Megan Kalomiris is a postbaccalaureate fellow in the Laboratory of Infectious Diseases (National Institute of Allergy and Infectious Diseases), where she studies noroviruses. After completing her training in 2021, she plans to pursue a master’s degree in science writing with hopes of working in science communications some day. In her spare time, she enjoys taking walks in the woods and playing games (now, virtually) with her friends.
Session 1: Tracking, Diagnostics, and Prevention
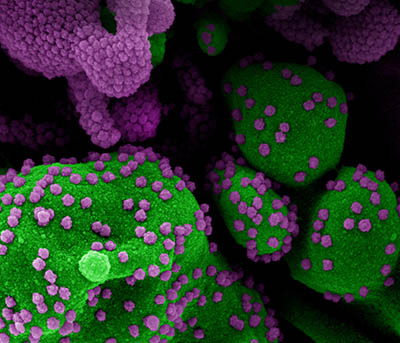
CREDIT: NIAID
Colorized scanning electron micrograph of an apoptotic cell (green) heavily infected with SARS-COV-2 virus particles (purple), isolated from a patient sample. Image at the NIAID Integrated Research Facility (IRF) in Fort Detrick, Maryland.
A half-dozen scientists described their research on tracking, diagnosing, and preventing COVID-19.
Stadtman Investigator Kaitlyn Sadtler (National Institute of Biomedical Imaging and Bioengineering) explained how the trans-NIH COVID-19 Serosurvey team mapped the pandemic with SARS-CoV-2 seropositivity. They used mail-in blood-sampling kits to detect antibodies to the SARS-CoV-2 spike protein among 10,000 donors, selected from a pool of 400,000 volunteers using integrated census data for a nationally representative sample. Preliminary results are expected to be published shortly, and the study is continuing with follow-up measurements through summer 2021.
Kaiyuan Sun, a postdoctoral fellow at the Fogarty International Center, talked about his research on transmission risk. His group analyzed data from Hunan, China, that evaluated 1,178 SARS-CoV0-2 infected individuals and their 15,648 close contacts. His team found that 80% of secondary transmissions can be traced back to 15% of SARS-CoV-2 infections and that a typical SARS-CoV-2 infection peaks just before the symptoms appear, with about 50% of the transmission occurring in the pre-symptomatic phase. The highest rate of transmission was household contact, followed by extended family, social or community, and health-care contact. Controlling the virus through nonpharmaceutical interventions requires case identification, isolation, and population-level intervention methods.
To introduce two talks on vaccine development, Barney Graham, the deputy director of the Vaccine Research Center at the National Institute of Allergy and Infectious Diseases (NIAID), explained how the mRNA-1273 vaccine rapidly controls SARS-CoV-2 infection in the upper and lower airways of nonhuman primates. Product development with Moderna, Inc., and clinical evaluation began in record time. The mRNA-1273 is in phase 3 clinical trials and was recently found to be 94% efficacious and received FDA’s Emergency Use Authorization approval on December 18, 2020.
Neeltje van Doremalen from NIAID’s Rocky Mountain Labs (Hamilton, Montana) presented preclinical studies of ChAdOx1 nCoV-19, an adenovirus-vectored vaccine that encodes the spike protein of SARS-CoV-2 and prevents pneumonia in rhesus macaques (Macaca mulatta). The vaccine was developed at the University of Oxford (Oxford, England) in partnership with AstraZeneca. Currently, phase 3 clinical trials are ongoing in the United Kingdom, the United States, Brazil, and South Africa.
Scientists have been unable to find specific immune-based biomarkers to predict clinical outcomes of COVID-19. NIAID investigators may be on to something, though, according to clinical infectious disease fellow Michael Abers. His team analyzed concentrations of 66 soluble biomarkers in 175 Italian patients with COVID-19 and identified 12 biomarkers that were significantly associated with mortality from SARS-CoV-2 infection. Four of them were consistently higher throughout the hospitalization in patients who died versus in those who recovered. If validated, the findings may offer the opportunity to develop personalized strategies for risk assessment and tailored treatments for hospitalized patients with severe COVID-19.
Yu Zhang (NIAID) presented findings from a recently published Science article regarding the genetic determinants of susceptibility to severe COVID-19 infection. (Science 370:eabd4570, 2020). “Disregarding preexisting conditions and age, a small proportion of patients were still at higher risk compared to others,” said Zhang. “This made us question if there is something in the host genome that affects severity.”
After an analysis of over 1,000 whole-genome sequencing and whole-exome sequencing of COVID-19 patients, Zhang and her colleagues from NIH and the COVID–Human Genetic Effort consortium determined that COVID-19 severity could be explained by mutations in type I interferon (IFN) response genes. Individuals who carry specific IFN deleterious variants can be more susceptible to infectious diseases.

Frances Fernando is a postbaccalaureate fellow in the Division of Intramural Population Health Research (Eunice Kennedy Shriver National Institute of Child Health and Human Development). After completing her NIH training in 2021, she plans to pursue a doctorate in public health to work on integrative global health issues and human rights. Outside of work, she gardens and explores Washington, D.C., by bike.
Session 2: Virology 1: Replication, Evolution, and Host Factors
“We are very much learning about COVID-19, but one area that we still lack is [understanding its] cell biology,” explained Nihal Altan-Bonnet, a senior investigator at the National Heart, Lung, and Blood Institute and one of eight investigators describing their research in session two of the COVID-19 Research Workshop. Although scientists understand much about how coronaviruses enter cells and replicate, they know little about how the viruses exit cells.
In her talk, Altan-Bonnet explained that it was once thought that beta-coronaviruses, such as SARS-CoV-2, exit cells via the secretory pathway. However, when her lab used the drug brefeldin A to block transport out of the cell via that pathway, she found that the viruses were still able to escape by taking advantage of lysosomes to aid in their egress. Lysosomes contain digestive enzymes and typically break down old cell parts, bacteria, and even viruses. Altan-Bonnet found that lysosomes of beta-coronavirus-infected cells suffer a profound loss of acidification, which may explain why SARS-CoV-2 can survive in the lysosome. These findings could provide insights into the cellular and immunological abnormalities observed in patients and suggest new therapeutic modalities.
Blake M. Warner, an assistant clinical investigator in the National Institute of Dental and Craniofacial Research, studies the role of saliva in the infection and transmission of SARS-CoV-2. Using such techniques as single-cell RNA sequencing and digital droplet polymerase chain reaction analysis, his lab determined that salivary epithelial clusters had high expression of angiotensin converting enzyme 2 (receptors that allow SARS-CoV-2 to enter cells), that salivary glands were infected with the virus in autopsy tissue samples from SARS-CoV-2 positive individuals, and that the virus can replicate within the salivary glands.
Interestingly, a high viral load in saliva correlates with loss of taste or smell along with other oral symptoms, indicating oral infection. The lab also found that mask use reduces expulsion of this potentially infectious saliva tenfold. Warner’s data identify tissues and cell types vulnerable to SARS-CoV-2 infection and highlight that the oral cavity plays a central role in infection, host response, and transmission.

CREDIT: YEN-TING TUNG, NCATS
NCATS scientist Emily M. Lee talked about how the 3D Tissue Bioprinting Laboratory is developing a variety of 3D platforms in which tissue equivalents can be generated. Above: Model of human small-airway epithelial tissue showing ACE2 receptors (yellow); ciliated cells (red); cytoskeleton (green); and nuclei (blue).
The session ended with a presentation by Emily M. Lee, a scientist in the 3D Tissue Bioprinting Laboratory at the National Center for Advancing Translational Sciences. To mimic the complexity of tissues in the human body, the lab is developing a variety of 3D platforms in which tissue equivalents can be generated including static 3D cultures as well as microfluidics 3D tissue devices that allow media or cells to flow through a system. The lab is creating 3D cellular lung models for studying the infectivity of high-impact respiratory viruses and the rapid testing of antiviral agents. These biofabricated tissues imitate tracheobronchial, small airway, and alveolar lung tissues in vitro and can be used to improve disease modeling, and for drug toxicology and efficacy studies.
Lee’s research uses a range of techniques including cell imaging, cytokine measurements, and single-cell RNA sequencing to identify the factors that determine whether a respiratory virus will cause severe or mild disease. Moreover, this data may help uncover new drug targets in host pathways. “We are utilizing and developing [these tissue model platforms] not only for SARS-CoV-2, but hopefully in preparation for the next disease X,” said Lee.

Natalie Hagen is a postbaccalaureate research fellow in the National Center for Advancing Translational Sciences, where she is performing pharmacokinetics studies of novel drug candidates. After her fellowship is over in 2021, she is planning to pursue a Ph.D. In her spare time, she likes to run, read, and go hiking with friends.
Session 3: Virology II: Structure, Function, and Inhibition of Viral Proteins
A dozen scientists presented their research on trying to understand SARS-CoV-2 infections by investigating the structure, function, and inhibition of viral proteins. Of particular interest was the spike protein that binds to the human receptor ACE2, which enables SARS-CoV-2 to enter target cells. The spike protein is of critical importance to the development of treatments and vaccines.
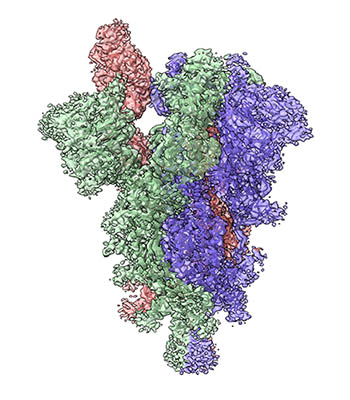
CREDIT: TONGQING ZHOU, NIAID
The spike protein on the surface of the SARS-CoV-2 virus allows binding to receptors on the host cell and is the major component of many COVID-19 vaccines. Shown: Cryogenic electron microscopy image of the structure of the SARS-CoV-2 spike with three subunits colored green, blue, and pink. The receptor-binding domain in the pink subunit is shown in the “up” position ready for binding to the receptor.
Presenters included Dominic Esposito (Frederick National Lab), who is optimizing the production of large amounts of spike proteins to supply to NIH scientists investigating the structure of the SARS-CoV-2 virus. Staff scientist Tongqing Zhou (National Institute of Allergy and Infectious Diseases, NIAID) is exploring how the virus uses pH changes along the endosomal pathway to bind to ACE2 and evade immune surveillance. Adam Olia’s (NIAID) finding that the spike protein unfolds slowly at neutral pH and rapidly refolds at low pH may have implications for spike-based vaccines. Senior investigator Mitchell Ho (National Cancer Institute), whose lab specializes in isolating single-domain antibodies (also called nanobodies) from camels, presented data showing three nanobodies that effectively inhibited interaction between SARS-CoV-2 and ACE2. Venkata Dandey from Mario Borgnia’s lab (National Institute of Environmental Health Sciences) also presented data on four humanized nanobodies that displayed varying degrees of potency in blocking virus interaction with ACE2. Because of their small size, nanobodies can be used in treatments, especially ones administered by an inhaler.
An interesting study from the lab of Peter D. Sun (NIAID), presented by staff scientist Jinghua Lu, showed how binding SARS-CoV-2 to ACE2 enzymes increased enzymatic activity. These results shed new light on the pathogenesis of COVID-19 and its complications and may lead to better treatments.
Nadya Tarasova (NCI) described how she used the Biowulf supercomputer to run virtual screens on synthetic compound libraries containing upward of a billion different structures; 190 hits were verified as SARS-CoV-2 protein ligands that are promising drug candidates and powerful chemical biology tools. The other presenters also described their efforts to use structural biology to unlock the mysteries of SARS-CoV-2 and how to fight it.

Sunita Chopra is a visiting postdoctoral fellow in the National Cancer Institute’s Radiation Oncology Branch, where she studies radiation-responsive coding and noncoding RNA signatures in the blood of whole-body-irradiated animal models. She plans to transition into a writing-intensive career after finishing her training at NIH. Outside of work, she enjoys being outdoors, cooking, listening to Bollywood songs, and being with friends.
Session 4: Clinical Manifestations and Pathogenesis
Eleven scientists gave short presentations highlighting what’s known so far about the clinical manifestations and pathogenesis of COVID-19. Following are descriptions of a few of the talks.
Temesgen Andargie, a research fellow in the National Heart, Lung, and Blood Institute’s (NHLBI’s) Laboratory of Applied Precision Omics, discussed his research demonstrating that cell-free DNA (cfDNA), which is released into the bloodstream by damaged cells, can help predict which hospitalized COVID-19 patients will need to go to the intensive care unit (ICU) and who might eventually die. Previous research has shown that cfDNA is detected in lung-transplant recipients whose bodies reject the organ, but before they develop symptoms. Andargie’s group found that in hospitalized COVID-19 patients, ICU patients have a sixfold increase of cfDNA from different tissue types in their blood compared with non-ICU patients. They also found that at admission, cfDNA is strikingly increased in COVID-19 patients who eventually die compared with those who recover. The research suggests cfDNA might be a useful biomarker to predict clinical trajectories and patient outcomes.
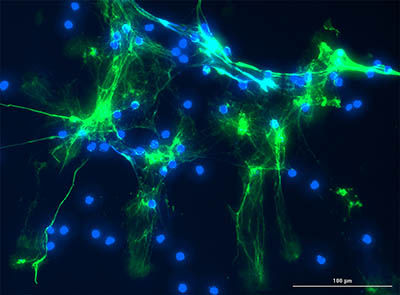
CREDIT: YOGEN KANTHI (NHLBI) AND JASON KNIGHT LAB (U MICHIGAN)
Yogen Kanthi (NHLBI) discussed his work, in Session 4, on how neutrophils are hyperactivated during a COVID-19 infection. Shown: Serum from patients with COVID-19 triggers healthy human neutrophils to undergo NETosis (green-citrullinated histone H3, a marker of NETosis; blue-DNA).
Lasker Clinical Research Scholar Yogen Kanthi (NHLBI) discussed his work on how neutrophils (a type of immune cell) are hyperactivated during a COVID-19 infection. When neutrophils die, they release neutrophil extracellular traps (NETs), bundles of sticky DNA that trap foreign microbes and can serve as building blocks for blood clotting. In COVID-19, hyperactivated neutrophils contribute to blood clots in the lung and lung damage. Kanthi’s group discovered that COVID-19 induces the production of prothrombotic autoantibodies that may contribute to NETs and blood clotting. They are interested in treating COVID-19 with dipyridamole, an FDA-approved medication that prevents blood clotting. Upon finding that dipyridamole inhibits neutrophil and NET formation, Kanthi launched a randomized, placebo-controlled trial to test the efficacy of the drug in patients with COVID-19.
Ben Afzali, a Stadtman Investigator in the National Institute of Diabetes and Digestive and Kidney Diseases, discussed how SARS-CoV2 activates the complement system (part of the immune response) in infected lung epithelial cells and how the complement proteins and COVID-19 together activate T cells in the lungs. In patients with severe COVID-19, the inflammatory response remains ramped up, changing the composition of RNA inside T cells and putting those T cells in a dangerous state of hyperactivation. Hyperactivated T cells contribute to the inflammation that causes lung damage in COVID-19 patients. Afzali’s group found that once activated, T cells switch on the vitamin D response pathway, which later initiates the shutdown of the immune response. People with a deficiency of vitamin D are more susceptible to contracting COVID-19 and also to developing more severe disease. The study data predicted that therapy with vitamin D, possibly in combination with steroids, may aid in the resolution of hyperinflammation in severe COVID-19.

Ethan Smith is a postbaccalaureate fellow in the National Institute of Nursing Research. He is working on clinical studies involving biomarkers for traumatic brain injury. Outside of work he enjoys playing board games with friends, reading, and watching television.
Session 5: Host Response and Immunology
“Optimism is the faith that leads to achievement. Nothing can be done without hope and confidence.” This quote by Helen Keller is the echo of the world today where we are fighting against an invisible enemy every day and every moment. Yes, we are living in a pandemic.
In the “Host Response and Immunology” session, 10 scientists discussed their work. Yvonne Baumer, a staff scientist in Tiffany Powell-Wiley’s lab (National Heart, Lung, and Blood Institute) talked about the impact of socioeconomic disparities and neighborhood deprivation (limited local resources of all types, financial and otherwise, that reflect structural inequalities) on COVID-19 severity and outcome. COVID-19 disproportionately affects African Americans and individuals residing in lower-resourced neighborhoods. Using in vivo and ex vivo approaches, Powell-Wiley’s team determined that the more disadvantaged neighborhoods (as measured by the Neighborhood Deprivation Index) people were from, the greater the decrease in certain toll-like receptors (TLRs), which are recognition molecules for multiple pathogens. A decrease in TLR2 in particular was associated with decreased expression of monocyte TLR2, a phenomenon associated with decreased survival in pneumonia. Further studies are needed to provide therapeutic insights.
Phillip Swanson (National Institute of Allergy and Infectious Diseases) talked about the efficacy of one of the messenger RNA (mRNA) vaccine candidates, mRNA-1273, which was codeveloped by Moderna, Inc., and NIAID’s Vaccine Research Center. The vaccine includes mRNA, encoding the full-length SARS-CoV-2 spike protein, encapsulated in a lipid nanoparticle that enables the mRNA to enter human cells. Swanson and his colleagues demonstrated that mRNA-1273 induces a strong T-cell helper type 1 (Th1) response in adults regardless of age and that the lack of Th2 responses indicated that the vaccine was safe to proceed to phase 3 testing. The vaccine has since gone on to phase 3 clinical trials and been found to be safe and 94% efficacious.
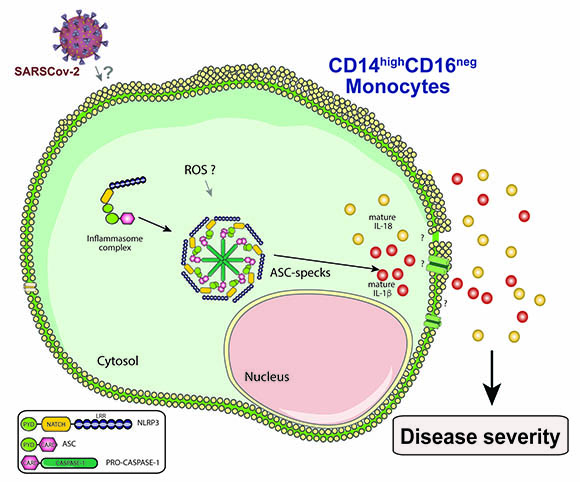
CREDIT: SILVIA L. LAGE AND EDUARDO P. AMARAL
Silvia Lucena Lage described how the NLRP3 inflammasome is activated in a type of monocyte, called CD14highCD16-, circulating in the blood of COVID-19 patients, seems to contribute to COVID-19-associated hyper inflammation and severity. (ROS=reactive oxygen species; ASC-specks=Apoptosis-associated speck-like protein.)
Silvia Lucena Lage (NIAID) presented her work on how the NLRP3 inflammasome is activated in a type of monocyte, called CD14highCD16-, circulating in the blood of COVID-19 patients. Her findings suggest that the activation of the NLRP3 inflammasome in circulating CD14highCD16- monocytes contributes to the severity of COVID-19-associated hyperinflammation and could potentially represent a target for host-directed therapies (therapies that interfere with host cell factors rather than acting directly on a pathogen) against COVID-19.

June Guha, a visiting fellow in NIAID, is investigating how nuclear factor NF-kappa-B signaling shapes the function of dendritic cells in Toxoplasma gondii infection. She was previously a visiting fellow in NCI (2015–2018) working on T-cell signaling. Outside the lab, she is interested in science writing and communications; is an editor on the Fellows Editorial Board; enjoys reading about scientific research in different fields; and has a special interest in astronomy.
Session 6: Animal Models and Vaccines
BY CHARLESICE HAWKINS, OITE
Animal models are invaluable for understanding the mechanisms of the immune response and for predicting how well treatments and vaccines will work in humans. Several scientists talked about the different kinds of animal models they are using in their COVID-19 research.
Emily Speranza, an independent research scholar in the National Institute of Allergy and Infectious Diseases (NIAID), explained how her team compared subgenomic RNA (sgRNA) from lung samples of African green monkeys (Chlorocebus aethiops) inoculated with either active SARS-CoV-2 or an irradiated inactive form of the virus. Speranza’s team found that replication of the virus in the lungs appears to occur at high levels only in pneumocytes (cells that line the inner surface of the lungs), while macrophages drive the inflammatory response. She went on to describe future aims to develop sgRNA testing assays that are more indicative of active infection.
Routes of SARS-CoV-2 transmission are also a major concern. Postdoctoral fellow Julia Port (NIAID Rocky Mountain Labs, NIAID RML), using a Syrian golden hamster (Mesocricetus auratus) model, found that intranasal and aerosol exposure—compared to oral or fomite (objects or surfaces contaminated with infectious particles) exposure—led to more severe COVID-19. Increased virus titers in the lungs and trachea were also observed with aerosol exposure; and airborne transmission was more efficient than fomite transmission. The findings suggest that the route of infection, possibly through differences in viral deposition throughout the respiratory tract, contributes to the large variability in the severity of COVID-19.
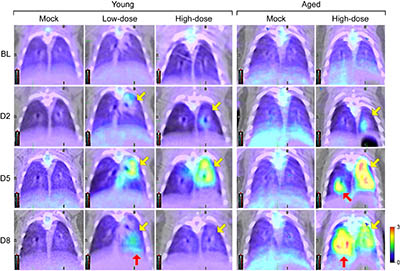
CREDIT: JI HYUN LEE AND JIRO WADA, IRF-FREDERICK, NIAID
NIAID researcher, Ji Hyun Lee, is using computed tomography (CT) and positron emission tomography (PET) imaging to characterize SARS-COV-2 in the golden hamster model. Shown: Fluorodeoxyglucose (FDG) PET images of lungs of young (4 to 5 weeks old) and aged (1 year old) hamsters that have been intranasally exposed to varying amounts of SARS-CoV-2. From top to bottom, images show how the disease progresses (from baseline to Day 8). The light green and yellow represent increasing metabolic activity in infected areas; infection peaked in young hamsters on Day 5 and resolved on Day 8, while aged hamsters showed mixed responses on Day 8 (yellow and red arrows).
Another NIAID researcher, Ji Hyun Lee, is using computed tomography (CT) and positron emission tomography (PET) imaging to characterize SARS-COV-2 in the golden hamster model. Radiological assessments by CT and PET imaging will allow real-time qualitative and quantitative serial monitoring of respiratory-tract changes in COVID-19 and enhance the evaluation of medical countermeasures. Also using the golden hamster model is FDA scientist Prabhuanand Selvaraj, who found that SARS-CoV-2 infection induces protective immunity that prevents re-exposure and limits transmission. Another FDA scientist, Meseda Clement, is using both mouse (Mus musculus) and hamster models to facilitate vaccine development by quantifying the immunogenicity of different forms of SARS-CoV-2 spike proteins.
Mice are also being used to study COVID-19, especially the transgenic K18-hACE2 mouse model, which expresses the human angiotensin-converting enzyme 2 receptor, the protein used by SARS-CoV-2 to enter cells. Lydia Roberts (NIAID RML), found that CD4+ and CD8+ T cells in K18-hACE2 mice display unique antigen specificity depending on their location in the lung. Tissue-resident T cells are directed against the nucleocapsid protein, whereas blood T cells are directed against the spike protein. Her findings may have important implications for future vaccines. With the same mouse model, Forrest Jessop (NIAID RML) used imaging to demonstrate metabolic perturbations among infected animals and characterized how SARS-CoV-2 manipulates the host metabolism. His findings will reveal targets that represent novel treatment strategies for COVID-19.
To determine whether genetic features of the host contribute to the severity of SARS-CoV-2 infection, staff scientist Shelly Robertson (NIAID RML) is working with Jackson Laboratories to assess disease susceptibility in a range of mouse strains crossed with the K18-hACE2 transgene. This approach has yielded models with varying degrees of disease severity and has demonstrated that the host’s genetic background influences the outcome of COVID-19.
Other innovative work is being carried out across NIH and FDA to improve vaccine efficacy, availability, and delivery. For example, NIAID staff scientist Baoshan Zhang, working with mice, is developing rapid and efficient vaccine platforms that use self-assembling nanoparticle immunogens to stimulate strong immune responses and provide protection against SARS-CoV-2. (Sci Rep 10:article number 18149, 2020)

Charlesice Hawkins, an administrative trainee in the Office of Intramural Training and Education, is pursuing a career in communications while also exploring her interests in creative writing and other forms of expression.
This page was last updated on Thursday, March 10, 2022
