Research Briefs
NHLBI: NEW VIRAL VECTOR FOR IMPROVED GENE THERAPY IN SICKLE-CELL DISEASE
CREDIT: NHLBI
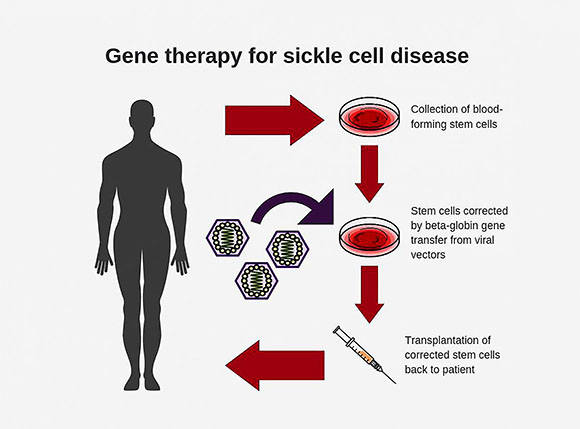
Diagram shows steps involved in conducting gene therapy for sickle-cell disease.
Researchers at NHLBI have developed a new and improved viral vector for use in gene therapy for sickle-cell disease. In advanced lab tests using animal models, the new vector was up to 10 times as efficient at incorporating corrective genes into bone-marrow stem cells as the conventional vectors currently used, and it had a carrying capacity of up to six times as high, the researchers report. The development of the vector could make gene therapy for sickle-cell disease much more effective and pave the way for wider use of it as a curative approach for the painful, life-threatening blood disorder. Sickle-cell disease affects about 100,000 people in the United States and millions worldwide.
Sickle-cell disease is an inherited blood disorder caused by a mutation, or misspelling, in the beta-globin gene. This mutation causes hemoglobin to produce sickle-shaped cells that can stick to the walls of blood vessels, causing blockage, pain, anemia, organ damage, and early death. With gene therapy, doctors modify the patient’s bone-marrow hematopoietic (blood-producing) stem cells in the lab by adding a normal copy of the beta-globin gene through the use of a viral vector. They then reinfuse the modified stem cells into the patient to produce normal, disc-shaped red blood cells.
For the past 30 years, researchers have been designing beta-globin vectors in a reverse structural orientation, meaning the therapeutic genes incorporated into the virus are translated, or “read,” from right to left by the viral vector-making machinery, much like reading an English sentence backwards. The process for making these vectors has been complicated, however.
About 10 years ago, John Tisdale and Naoya Uchida, a staff scientist in his lab, searched for an improved delivery vehicle and decided to undertake a radical redesign of the beta-globin vector. They created a new forward-oriented beta-globin vector that is read from left to right, making the gene translation approach less complicated.
The researchers tested the new vectors in mice and monkeys and compared the results with those from reverse-oriented vectors. They found that the new vectors could transfer a much higher viral load—up to six times as many therapeutic beta-globin genes than the conventional vectors—and had four to 10 times as much transduction efficiency, a measure of the ability to incorporate corrective genes into repopulating bone marrow cells. The new vectors also showed a capacity for longevity, remaining in place four years after transplantation. Researchers also found that they could be produced in much higher amounts than the conventional vectors, potentially saving time and lowering costs associated with large-scale vector production.
The new vector, for which the NIH holds the patent, still needs to undergo clinical testing in humans. Already an estimated 27 people with sickle-cell disease have undergone experimental gene therapy using conventional vectors. Through its Cure Sickle Cell Initiative [https://www.nhlbi.nih.gov/science/cure-sickle-cell-initiative], NIH is working to accelerate the development of these and other new genetic therapies, including gene editing, with the goal of finding a cure for the disease. (NIH authors: N. Uchida, M.M. Hsieh, L. Raines, J.J. Haro-Mora, S. Demirci, A.C. Bonifacino, A.E. Krouse, M.E. Metzger, R.E. Donahue, and J.F. Tisdale, Nat Commun 10:4479, 2019; DOI:10.1038/s41467-019-12456-3)
NINDS, NICHD: ALS GENE MAY BE A HITCHHIKER’S GUIDE TO THE NEURON
CREDIT: NINDS
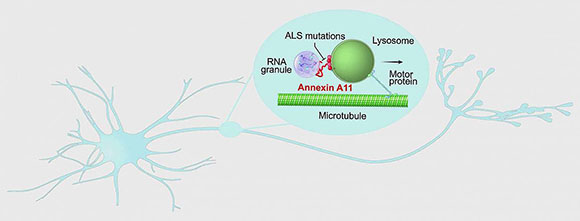
Researchers discovered that annexin A11 (encoded by the ANXA11 gene), which is linked to a rare form of amyotrophic lateral sclerosis (ALS), may play a critical role in the transport of RNA-encoded housekeeping instructions throughout neurons by hitching RNA granules to traveling lysosomes and that disease-causing mutations prevent hitchhiking.
Affecting at least 14,000 Americans, amyotrophic lateral sclerosis (ALS) is a paralyzing and highly fatal neurodegenerative disorder for which there are no effective treatments.
In a new experiment, scientists peered inside neurons and watched the workings of the protein annexin A11, encoded by the ANXA11 gene, which is linked to a rare form of ALS. Using advanced live-cell microscopy, they found that neurons may normally use the gene to ship internal housekeeping instructions via a newly discovered “hitchhiking” system and that disease-causing mutations may tie up deliveries at the cell’s loading docks.
The study, published in Cell, was led by researchers at NINDS, Howard Hughes Medical Institute Janelia Research Campus (Ashburn, Virginia), and the Cambridge Institute for Medical Research (Cambridge, England).
The scientists also found that disease-causing mutations in ANXA11 prevented hitchhiking, which in turn prevented RNA from being delivered to the far reaches of neurons. Many genetic studies have found that ALS is often caused by mutations in genes known to play roles in either RNA processing or the control of lysosomes. These results suggest that there is a link between these seemingly different processes and that understanding this type of hitchhiking in neurons may lead to new treatments for ALS. (NIH authors: M.S. Fernandopulle, L. Hao, R. Patel, M. Nelson, M.A. Gachechiladze, C.A. Stephens, L.R. Forrest, and M.E. Ward, Cell 179:147–164.e20, 2019; DOI:10.1016/j.cell.2019.08.050)
NIAID (ROCKY MOUNTAIN): HOUSEHOLD BLEACH INACTIVATES CHRONIC WASTING DISEASE PRIONS
A five-minute soak in a 40% solution of household bleach decontaminated stainless steel wires coated with chronic wasting disease (CWD) prions, according to a new study by NIAID scientists at the Rocky Mountain Laboratories in Hamilton, Montana. The scientists used the wires to model knives and saws that hunters and meat processors use when handling deer, elk, and moose—all of which are susceptible to CWD, a brain-damaging and fatal prion disease in cervids (members of the deer family).
To date, CWD has not been found in people. However, other prion diseases can affect people, therefore scientists, wildlife managers, and public-health agencies have suggested handling CWD cervid tissues with caution. CWD is spreading in North America, increasing the potential for human exposure. The disease has been found in cervids in 26 states and three Canadian provinces, as well as in Norway, Finland, and South Korea. Not all animals infected with CWD will show signs of disease, but those that do appear weak and thin.
Infectious prions are extremely difficult to inactivate, which led the scientists to seek a practical, low-cost CWD decontamination method. Bleach has been proven as a decontaminant against other types of prions but had never been tested against CWD. CWD prions adhere readily to stainless steel and can contaminate knives, saws, and other equipment.
The researchers worked with CWD-infected brains from white-tailed deer (Odocoileus virginianus) and mule deer (O. hemionus). They tested various bleach concentrations and soak times to determine the most-effective combination to eliminate prion seeding. Notably, the study failed to find an effective method to decontaminate CWD-infected solid tissue. Pieces of CWD-infected brain retained prion activity even after a 30-minute soak in 100% bleach. Investigators note that bleach fails to penetrate tissues and should be used only as a surface decontaminant.
The scientists hope that public-health and wildlife agencies will consider this study when making formal recommendations for decontamination of CWD prions. (NIH authors: K. Williams, A.G. Hughson, B. Chesebro, and B. Race, PLOS One 14:e0223659, 2019; DOI:10.1371/journal.pone.0223659)
NCCIH: STUDY IN MICE EXPLAINS HOW BRAIN CAN TURN PAIN SIGNALS UP OR DOWN
CREDIT NCCIH
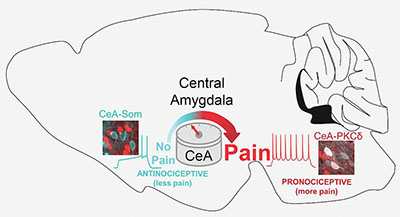
The central amygdala (CeA) functions as a pain rheostat in the forebrain, enhancing or attenuating pain. Modulation of pain is accomplished by cell-type-specific changes in activity after injury. Activity in CeA neurons expressing protein kinase C, delta type (PKCδ; right) drives increases in pain-related behaviors whereas activity in CeA neurons expressing somatostatin (CeA-Som; left) reduces pain-like behaviors.
A new NCCIH study in mice uncovered a previously unknown role that the central amygdala can play in upgrading or downgrading pain signals in the brain’s circuitry. “Early research showed that the central amygdala, long known for its role in processing fear, can dial up pain signals. Yet other studies have pointed to the central amygdala’s role in suppressing pain or prompting an analgesic response,” said Yarimar Carrasquillo, senior author of the study, which appeared in Cell Reports. “This study unravels what seemed to be a contradiction in early research and reveals a previously hidden ‘switch’ in the central amygdala that can turn up or turn down pain signals.” (NIH authors: T.D. Wilson, S. Valdivia, A. Khan, H.S. Ahn, A.P. Adke, S.M. Gonzalez, Y.K. Sugimura, and Y. Carrasquillo, Cell Rep 8:332-346.e5, 2019; DOI:10.1016/j.celrep.2019.09.011)
OD, NIGMS: CHOICE OF RESEARCH TOPIC CONTRIBUTES TO PERSISTENT GAP IN NIH RESEARCH GRANTS TO BLACK SCIENTISTS
Research-topic preference accounts for more than 20% of a persistent funding gap for black scientists applying for NIH research project (R01) grants compared with white scientists, according to a new study by NIH scientists. Researchers examined each step in the application-submission and -review process for R01 applications submitted between 2011 and 2015. The study confirms previous findings that career stage and institutional resources influence the gap in the number of submissions by black and white researchers. However, the finding that black applicants as a group are more likely to propose research topics that are less likely to be funded was new.
Black applicants were more likely than white applicants to propose research topics that receive awards at a lower rate, such as community- or population-level research. Research topics with the lowest success rates included health-disparities research and patient-focused interventions. Importantly, white researchers also experienced lower award rates in these topic areas, although less so than black researchers.
Despite significant efforts by NIH to close this funding gap, first identified in 2011, it continues to persist. This new research identifies opportunities for interventions that could bolster NIH’s scientific workforce diversity efforts underway, such as the Diversity Program Consortium and the Maximizing Opportunities for Scientific and Academic Independent Careers (MOSAIC; K99/R00 and UE5) program.
“The underlying explanation for the phenomenon observed in this paper is complex and requires further study to determine how best to intervene,” said Hannah A. Valantine, NIH’s Chief Officer for Scientific Workforce Diversity, and one of the authors of the study. “We will continue to explore the causal factors behind the R01 funding gap to add to NIH’s broader scientific workforce diversity program.” (NIH authors: T.A. Hoppe, A. Litovitz, K.A. Willis, R.A. Meseroll, M.J. Perkins, B.I. Hutchins, A.F. Davis, M.S. Lauer, H. A. Valantine, J.M. Anderson, and G.M. Santangelo, Science Advances 5:eaaw7238, 2019; DOI:10.1126/sciadv.aaw7238)
NINDS, NICHD, NIDCD, NHLBI: “STICKY” GENE MAY HELP VALIUM CALM NERVES
CREDIT: WEI LU LAB, NIH/NINDS
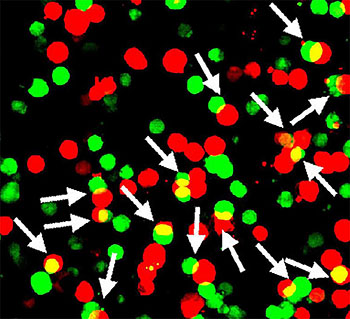
In a study of mice, NIH researchers showed that a protein encoded by a gene called Shisa7 (green) may boost the nerve-calming effects of diazepam (a.k.a. Valium) and other benzodiazepines by sticking to GABA type A neurotransmitter receptors (red).
Between 1999 and 2017, the United States experienced a tenfold increase in the number of people who died from overdoses of diazepam (a.k.a. Valium) and other benzodiazepines. For years, scientists thought that these powerful sedatives, which are used to treat anxiety, muscle spasms, and sleeping disorders, worked alone to calm nerves. Now, in an article published in Science, NIH researchers show that this view of the drugs and the neural circuits they affect may have to change. In a study of mice, scientists discovered that both may need the assistance of a “sticky” gene, named after a mythological figure, called Shisa7.
“We found that Shisa7 plays a critical role in the regulation of inhibitory neural circuits and the sedative effects some benzodiazepines have on circuit activity,” said NINDS Stadtman Investigator Wei Lu, the senior author of the study. “We hope the results will help researchers design more-effective treatments for a variety of neurological and neuropsychiatric disorders that are caused by problems with these circuits.”
Before this study, it was thought that benzodiazepines worked alone to boost the nerve-calming responses of GABA-A receptors. Lu’s lab found that, instead, these responses may depend greatly on whether a protein encoded by the Shisa7 gene is stuck to GABAA receptors.
Like many scientists, Lu thought that Shisa7 played a role in controlling a completely different type of synapse that relies on the neurotransmitter glutamate to excite, rather than quiet, neurons. Recent studies suggested that Shisa7 along with other Shisa genes encodes proteins that adhere to glutamate receptors. Once attached, these “auxiliary” proteins can control a receptor’s response to glutamate or its presence at synapses. Then, a few years ago, Lu’s team noticed something interesting in a scientific article on the shisa proteins. “We found the results striking. The paper showed that Shisa7 was the only protein in this family that seemed to have no effect on the activity of an important type of glutamate receptor,” said Lu. “This caught our attention and we decided to take a closer look.”
The researchers in Lu’s lab systematically examined Shisa proteins in mouse neurons. To their surprise, they found that Shisa7 appeared to play a unique and critical role in the nerve quieting GABA synapses.
“Our results shine a spotlight on the potential clinical importance of auxiliary proteins like shisa 7,” said Lu. “Many of the neurological drugs we use today are designed to control the activity of synaptic receptors. For the first time, we show that researchers may also want to consider auxiliary proteins like shisa 7 in developing new treatments that target GABA-A receptors.”
His team plans to explore in greater detail the role Shisa7 may play in inhibitory circuits and in other neurological disorders. (NIH authors: W. Han, J. Li, K.A. Pelkey, S. Pandey, Y.-X. Wang, K. Wu, L. Ge, T. Li, D. Castellano, C. Liu, L.-G. Wu, R.S. Petralia, C.J. McBain, and W. Lu; Science 366:246–250, 2019; DOI:10.1126/science.aax5719)
NIAID (ROCKY MOUNTAIN LABS): NIH SCIENTISTS DEVELOP TEST FOR UNCOMMON BRAIN DISEASES
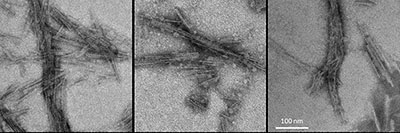
CREDIT: NIAID
Representative negative-stained transmission electron microscopy images of 4-repeat tau protein amplified in a real-time, quaking-induced conversion process (4R RT-QuIC) products seeded with brain homogenates from individuals with the designated diseases: frontotemporal dementia and Parkinsonism linked to chromosome 17; corticobasal degeneration; and progressive supranuclear palsy.
NIAID scientists at Rocky Mountain Laboratories (Hamilton, Montana) have developed an ultrasensitive new test to detect abnormal forms of the protein tau associated with uncommon types of neurodegenerative diseases called tauopathies. This advance gives them hope of using cerebrospinal fluid (CSF) to diagnose these and perhaps other, more common neurological diseases, such as Alzheimer disease.
The abnormal deposition of tau in the brain is linked to at least 25 different neurodegenerative disease, but to accurately diagnose them, researchers must often analyze brain tissue after the patient has died. For their study, the researchers used the same test concept they developed when using post mortem brain tissue samples to detect the abnormal tau types associated with Pick disease, Alzheimer disease, and chronic traumatic encephalopathy. They adapted the test to use CSF for the detection of abnormal tau of progressive supranuclear palsy, corticobasal degeneration, and other less common tauopathies.
They detected abnormal tau in CSF from both living and deceased patients. The new test is called 4R RT-QuIC, which stands for 4-repeat tau protein amplified in a real-time, quaking-induced conversion process. The researchers plan to continue evaluating the clinical performance of 4R RT-QuIC by analyzing larger sets of CSF samples. One focus will be to compare test results from tauopathy patients who agree to provide CSF samples both before and after death. The scientists hope this type of evaluation will help them better understand how abnormal tau in CSF evolves during brain disease. (NIH authors: E Saijo, M.A. Metrick 2nd, A. Kraus, and B. Caughey, Acta Neuropathol, 2019; DOI:10.1007/s00401-019-02080-2)
NINDS: MICROBLEEDS MAY WORSEN OUTCOME AFTER HEAD INJURY
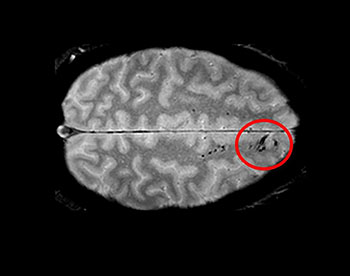
CREDIT: LATOUR LAB/NINDS
Traumatic microbleeds appear as dark lesions on MRI scans and suggest damage to brain blood vessels after head injury.
Using advanced imaging, NINDS researchers have uncovered new information about traumatic microbleeds, which appear as small, dark lesions on magnetic-resonance imaging (MRI) scans after head injury but are typically too small to be detected on computed tomography (CT) scans. The findings suggest that traumatic microbleeds are a form of injury to brain blood vessels and may predict worse outcomes.
The researchers, who also included researchers from Cold Spring Harbor Laboratory (Cold Spring Harbor, New York) and the Uniformed Services University of the Health Sciences (Bethesda, Maryland), studied 439 adults who experienced head injury and were treated in the emergency department. The subjects underwent MRI scans within 48 hours of injury and again during four subsequent visits. Participants also completed behavioral and outcome questionnaires.
The results showed that 31% of all study participants had evidence of microbleeds on their brain scans. More than half (58%) of participants with severe head injury showed microbleeds as did 27% of mild cases. Most patients who exhibited microbleeds had both types. The findings also revealed that the brain’s frontal lobes were most likely to show microbleeds and that patients with microbleeds were more likely to have greater disability than patients without microbleeds.
The family of a participant who died after completion of the study donated the brain for further analysis. The research team imaged the brain with a more powerful MRI scanner and conducted detailed histological analysis. The results showed iron, indicating blood, in macrophages tracking along the vessels seen on the initial MRI as well as in extended areas beyond that seen on MRI.
The authors note that microbleeds after brain injury may be a potential biomarker for identifying which patients may be candidates for treatments that target vascular injury. More research is needed, however, to determine additional effects of microbleeds and ways to treat them. There is currently no evidence that MRI scans should replace CT scans for suspected head injury. (NIH authors: A.D. Griffin, L.C. Turtzo, Z. Lodato, A.D. Moses, G. Nair, D.P. Perl, N.A. Edwards, B.J. Dardzinski, R.C. Armstrong, A. Ray-Chaudhury, and L.L. Latour, Brain, awz290, 2019; DOI:10.1093/brain/awz290)
This page was last updated on Wednesday, March 30, 2022
