Understanding the Body’s Universe of Atoms
IRP’s Adriaan Bax Elected to Royal Society for Pioneering Nuclear Magnetic Resonance Methods

IRP Distinguished Investigator Adriaan Bax was elected a Fellow of the Royal Society earlier this year for his contributions to a scientific technique that sheds light on the structure and function of molecules.
“My mortal body is indeed a universe of atoms, but I am just an atom in the universe myself,” physicist Richard Feynman once wrote in a poem. Within our own bodily universes, all those atoms tell a story about how our bodies work — and how they sometimes don’t.
IRP Distinguished Investigator Adriaan Bax, Ph.D., has pioneered ways to watch those atoms using nuclear magnetic resonance (NMR) methods. This year, he was elected a Fellow of the Royal Society in recognition of his research, which has contributed greatly to our knowledge of how the proteins and nucleic acids that keep our bodies running smoothly are structured, how they move, and how they interact, particularly in relation to diseases caused by their malformations and malfunctions. The Royal Society, which was founded in England in the 1660s, is a fellowship of many of the world’s most eminent scientists and is the oldest scientific academy in continuous existence.
“It is, of course, very exciting for me to receive this exceptional honor,” Dr. Bax says. “I am particularly pleased because I still feel a strong connection with Great Britain, where I received much of my graduate student training and where I became really excited about pursuing a career in research.”
Every molecule in existence is made up of individual atoms of the various elements in the periodic table, so marking and tracking atoms within molecules can yield important insights into their structure and movement. Dr. Bax’s specialty, NMR spectroscopy, measures very weak radiofrequency signals emitted by certain atoms when they’re placed in a strong magnetic field. Dr. Bax specifically focuses on creating ways to link each radiofrequency signal to a specific atom in the molecule, which allows for a more extensive and in-depth analysis and visualization of molecules — a vital topic of study given that those molecules can make the difference between a healthy and unhealthy body.

An NMR spectrometer
“My work has focused on improving the technology to make it more sensitive and to allow us to study more complex systems,” Dr. Bax says. “There is much practical interest in our work, both for pharmaceutical applications to develop new medicines and for basic research to gain deeper insights into how proteins move and interact with one another.”
Most people have heard of magnetic resonance imaging (MRI), which detects signals from the hydrogen atoms in the body's water molecules and uses them to create a map of organs, empty spaces, and abnormalities. NMR spectroscopy is conceptually similar to MRI, except that it focuses on small samples of specific molecules, including proteins, and detects signals from a range of atoms, such as hydrogen, carbon, nitrogen, and phosphorus. The way those atoms are arranged in a molecule determines its shape, and that shape, in turn, determines its function. Proteins can also adjust their shape to better interact with partners or in response to binding with a drug. Problems can occur when proteins fold into abnormal shapes, causing them to stick together in specific ways. Diseases like Alzheimer’s and Parkinson’s, which are characterized by a buildup of amyloid plaques and hardened bits of other proteins, are examples of the types of ‘protein-misfolding’ diseases that Dr. Bax studies using NMR spectroscopy.
“At their very basic level, many diseases occur when interactions between proteins and other molecules in our bodies fail to function properly,” Dr. Bax explains. “Many diseases are related to a misfunctioning protein pathway.”
While NMR has a long history of delivering significant improvements on researchers’ abilities to visualize proteins, Dr. Bax’s primary contribution, triple resonance NMR, has expanded the scope of biological NMR considerably. In addition to the traditionally studied hydrogen atoms, his triple resonance NMR relies on carbon and nitrogen atoms containing an atypical number of neutrons in their nucleus, known as isotopes.
Dr. Bax’s team genetically modifies cells to make the proteins the group wants to study and feeds them a mixture containing high levels of those naturally occurring isotopes. The cells then incorporate the isotopes into the proteins they generate, initially as a linear string of chemically bonded building blocks called amino acids. The isotopes emit weak radiofrequency signals when exposed to the magnetic field in an NMR spectroscopy machine, allowing for their location in the protein to be mapped and followed as the linear, flexible string folds into a well-defined but partially flexible three-dimensional structure.
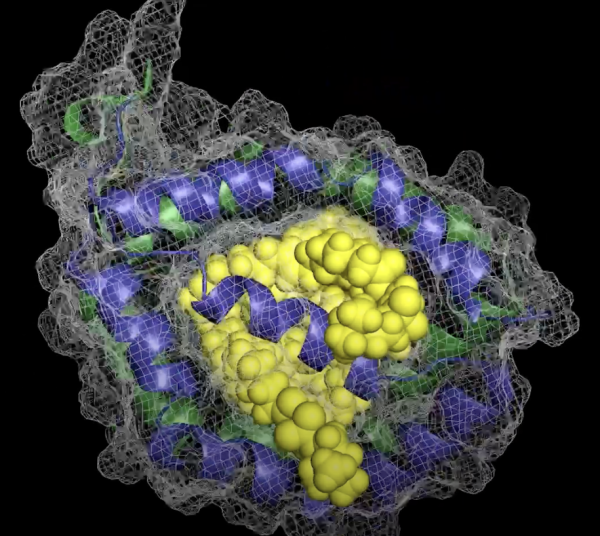
NMR can reveal detailed information about the shape of proteins, which determines how they function.
“Proteins are not rigid Lego blocks; they have built-in flexibility, and that flexibility is important for their function,” Dr. Bax explains. “We can measure this flexibility in great detail with NMR spectroscopy using signals from nitrogen isotopes.” For example, his team has demonstrated that a protein called calmodulin, found in the ‘smooth’ muscle that makes up many of our internal organs, binds to well over a hundred different targets, and it can do this, Dr. Bax adds, “because it has two sections, each of which behaves like a human hand, with the two of them tightly clasping onto their target from opposite sides.”1,2
“When we first tried to publish our development of triple resonance NMR, it was rejected by scientific journals with the argument that it was ‘technologically interesting, but we don’t need this kind of complicated technology,’” Dr. Bax recalls. “Now, it is the method of choice world-wide for studying proteins and their motions.”
Another important advancement Dr. Bax’s team made involves finding ways to ‘weakly orient’ proteins in a magnetic field. Molecules scientists want to study with NMR spectroscopy can be coached to all point in the same direction, like soldiers ready to march, by dissolving them in a liquid crystalline solvent, similar to the substance used in digital watch displays, then exposing them to a magnetic field. However, with large molecules, this uniformity causes NMR signals to become broad, fuzzy, and overly complex, making it impossible to see the signals clearly.

Creating new ways to study molecules with NMR sometimes means getting hands-on with the machines that perform it.
It’s a problem Dr. Bax had been working to correct since his undergraduate days in The Netherlands, and he finally solved it in 1997 using insights from a lecture on chemical techniques used in the petrochemical industry. The resulting approach, which uses a different substance to dissolve molecules, yields exceptionally clean information about the orientation of chemical bonds in a protein or a nucleic acid. This approach allows researchers to get a more accurate picture of molecular structure and increases the size of the molecules they can look at. The discovery, published in the prestigious journal Science, has become the cornerstone of numerous subsequent studies. 3,4
“This very precise information allows us to learn something about molecules’ motions, too,” Dr. Bax explains. “So, for example, if a drug binds on one side of the protein, does that impact the opposite side of the protein where, for example, a site that binds to other molecules might be located? That is really a gold mine of information because it yields insights into how signals can be transmitted in the body.”
Dr. Bax emphasizes that working at NIH has allowed him the freedom to pursue a lot of out-of-the-box ideas that had a high risk of failure. As a result, he has managed to make important discoveries that sometimes defy the expectations of his fellow scientists.
“I've got about 20 scientific papers that have been cited over a thousand times each, and nearly half of those were originally rejected for publication by peer reviewers,” he says. “The ones with the most impact were the hardest to get published.”
Subscribe to our weekly newsletter to stay up-to-date on the latest breakthroughs in the NIH Intramural Research Program.
References:
[1] Ikura M; Kay LE; Bax A. A novel approach for sequential assignment of 1H, 13C, and 15N spectra of proteins: heteronuclear triple-resonance three-dimensional NMR spectroscopy. Application to calmodulin. Biochemistry. 1990 Oct. 1; 29 (19): 4659–67. doi:10.1021/bi00471a022.
[2] Kay LE, Ikura M, Tschudin R, Bax A. Three-dimensional triple-resonance NMR spectroscopy of isotopically enriched proteins. J Magn. Reson. 1990 Oct. 1; 89 (3): 496–514. doi: 10.1016/0022-2364(90)90333-5.
[3] Tjandra N, Bax A. Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science. 1997 Nov. 7; 278(5340):1111-1114. doi: 10.1126/science.279.5340.1111.
[4] Bax A. Weak alignment offers new NMR opportunities to study protein structure and dynamics. Protein Sci. 2003; 12(1):1-16. doi: 10.1110/ps.0233303.
Related Blog Posts
This page was last updated on Wednesday, September 18, 2024
