For the Love of PCR
I hate PCR—the method we use in the lab to make millions of copies of a specific piece of DNA. You mix everything together that’s necessary for the reaction, and half the time it works, half the time it doesn’t. One day you get great PCR results, you’re on cloud nine, everything worked, and then you go repeat it to verify the result (because n never equals 1 in science), and it doesn’t work. You begin to feel like maybe you just got lucky with the first experiment. Things need to be reproducible in science so it’s tear-your-hair-out frustrating when things work one day and don’t the next.
But, this is science (and your hair looks much better on your head), and it’s not like anything you see on CSI where they get results in seconds and it works every time. The further along you get in your scientific career, the better you are at troubleshooting. That’s the next step when an experiment doesn’t work consistently, so you take a look at your primers, maybe design some new ones that might work better (or order some that are guaranteed by a company to work). Or you do an experiment with different annealing temperatures to make sure you’re using the optimal temperature. There are many things you can troubleshoot in a PCR reaction.
Maybe I’m getting ahead of myself. For those of you not quite as familiar with PCR, let me explain. PCR stands for polymerase chain reaction. Polymerase comes from the fact that DNA is a polymer, a very long chain of repeating units, and the “ase” ending indicates an enzyme, a type of protein that catalyzes a reaction. The reaction in PCR is extending DNA, using a single strand of DNA as a template and a primer to get the process started. Cells use polymerase to replicate their DNA before undergoing mitosis (splitting), so that each new cell has an identical set of genes. In the case of PCR, we’re only interested in one or more small areas of DNA, instead of the whole genome, and we make as many copies as possible rather than just one extra copy.
There are many different ways PCR is used. On the aforementioned CSI, the actors often use it to amplify several areas of a victim’s or suspect’s genome and compare the sequences to other samples to see if they match. I’m using PCR to confirm that I have “knocked down” proteins. In my experiments, small interfering RNA (siRNA) is tranfected (introduced) into the cell where it targets messenger RNA (mRNA), which is the precursor to protein. The expected result of siRNA transfection is that less of the protein gets made in the cells containing siRNA. The goal of the experiment is to confirm that and then determine what the impact was on the cell. However, before I can do any type of PCR, I have to take the mRNA from the cells and then make DNA. The process requires reverse transcription (RT), wherein mRNA serves as a template to create complementary DNA (cDNA).
Once I have cDNA, I can continue on and do PCR, and I use a specific type of PCR called quantitative PCR (qPCR) that allows smaller amounts of the product I’m looking for to be detected in real time. In a standard PCR, you can only detect the product after the reaction is over, and you have to perform the additional step of running the PCR products on a gel. With qPCR, the amount of DNA is measured using a fluorescent reporter molecule after the end of each PCR cycle. If the cDNA is abundant, the fluorescent signal from the reporter increases rapidly and reaches saturation during an early cycle of the PCR. If the cDNA is not so abundant, the fluorescent signal stays low longer and doesn’t increase until a much later cycle in the PCR, as you can see in the following figure.
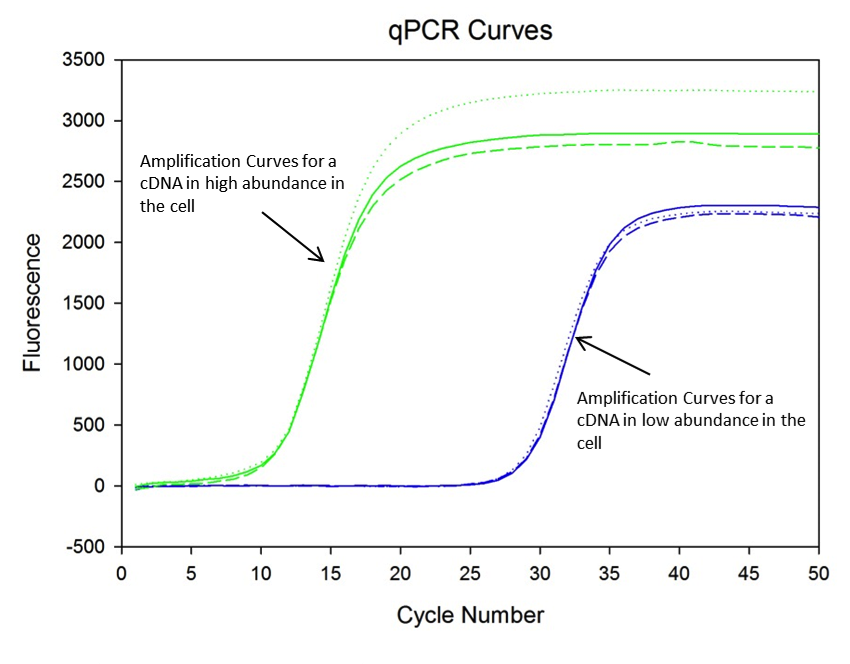
Once I get the qPCR results, I compare the siRNA knockdown sample to the untransfected sample and determine if there was less of the cDNA for the protein I’m looking for in the knockdown sample. Ideally, I would see the siRNA knockdown reaction’s fluorescence signal increase later than in the untransfected sample (the curves would be further to the right). Keep your fingers crossed that I will be able to get some awesome results to share with you in the near future!
Related Blog Posts
- IRP’s Thomas Kunkel Elected to National Academy of Sciences
- Tiny Molecules Have Big Potential for Treating Eye Diseases
- Global Scientists Come Together at the National Institutes of Health
- RNA-Targeting Therapeutic Restores Protein Absent in Spinal Muscular Atrophy
- CRISPR Pioneer Jennifer Doudna Headlines NHGRI 25th Anniversary Celebration
This page was last updated on Monday, January 29, 2024
