CRISPR Pioneer Jennifer Doudna Headlines NHGRI 25th Anniversary Celebration
It seems like every day there is a new story in a prominent news outlet about the revolutionary gene-editing approach known as CRISPR/Cas9. What these reports often fail to mention is all the scientific discoveries that paved the way for that groundbreaking technology, including the key contributions of government scientists working in the Intramural Research Program of NIH’s National Human Genome Research Institute (NHGRI). Last week, the NHGRI IRP celebrated its 25th anniversary with a day-long symposium headlined by a keynote from the co-discoverer of CRISPR/Cas9, University of California, Berkeley professor Dr. Jennifer Doudna.

Dr. Jennifer Doudna, co-discoverer of CRISPR/Cas9, was the keynote speaker at a symposium commemorating the 25th anniversary of the Intramural Research Program at the National Human Genome Research Institute (NHGRI).
Photo credit: Ernesto Del Aguila, NHGRI
In 2012, Dr. Doudna and her collaborator Dr. Emmanuelle Charpentier published an astonishing paper that detailed how scientists could leverage the CRISPR/Cas9 system to cut strands of DNA at specific locations, allowing researchers to remove or add new DNA sequences as they see fit. Scientists around the world immediately realized the implications of this development: it provides a one-stop-shop to easily and reliably edit DNA.
But Dr. Dounda certainly did not start off thinking she would rock the field of gene editing to its core. As a young girl growing up in Hilo, Hawaii, she did not personally know a single scientist, and her first introduction to genetics came at age 14 via James Watson’s book The Double Helix. Reading the book, she recalled, was a formative experience.
“In science, you could ask a question as complicated as ‘what’s the chemical structure of the code of life?’ and actually come up with experiments to figure it out,” Dr. Doudna said during her keynote speech at the NHGRI event. “That kind of blew my mind.”
While Dr. Doudna began her scientific career investigating how DNA produces proteins that perform important functions in cells, she eventually realized the key role that DNA-like genetic material called RNA plays in this and many other vital processes. Her interest in RNA eventually led her to Dr. Jill Banfield, a researcher who studies microbial DNA sequences isolated from environmental samples in order to identify the microbes present and the kinds of viruses that infect them. In 2005, the pair discussed the recent discovery of CRISPR sequences in bacteria, an observation that intrigued scientists at the time because they appeared to include sequences of DNA from viruses. Dr. Banfield suggested to Dr. Doudna that bacteria might use their CRISPR sequences to create RNAs that help them fight off viruses.
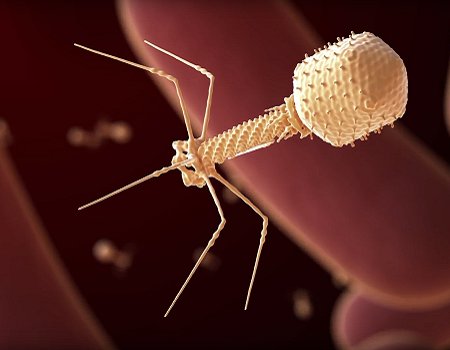
Viruses like this bacteriophage can infect and kill bacteria, forcing them to evolve systems like CRISPR/Cas9 that allow them to fend off viral assaults.
“In a curious way, what I thought was a very obscure area of research – something I thought was the most unusual thing I had spent time researching in my academic lab – turned into, eventually, a very powerful technology for manipulating genomes,” Dr. Doudna said.
Science is a world of acronyms, but even among the general word-salad of biology and chemistry, CRISPR/Cas9 stands out. The first half of this combo stands for Clustered Regularly Interspaced Short Palindromic Repeats, segments of microbial DNA that contain many instances of the same sequence of the DNA nucleotides A, C, T, and G. These repetitive sequences are palindromic, meaning they are identical no matter what direction you read them in. Each occurrence of these repeated sequences in a bacterium’s CRISPR regions is followed by a DNA sequence from a virus that has at some point infected that microbe.
Meanwhile, Cas genes like Cas9, which are found next to CRISPR sequences, encode Cas proteins. The CRISPR regions are used to create strands of RNA, which guide the Cas enzymes to the sequences of viral DNA found in the CRISPR regions, much like a seeing eye dog leads a blind man across the street. Because these guide RNA molecules were generated from the viral DNA sequences in the CRISPR regions, they can bind only to those specific DNA sequences or closely matching ones. When a Cas protein meets one of those viral DNA sequences, the guide RNA binds to it and the Cas enzyme cuts it up. Because viruses are little more than packages of DNA — most scientists don’t even consider them to be living organisms — the CRISPR/Cas duo allows microbes to defend themselves from viruses.
“It was amazing to us to think that bacteria had evolved this incredible protein: an RNA-guided enzyme whose job was to find and cut double-stranded DNA,” Dr. Doudna said, “but by understanding how it worked, we as scientists could then turn that around and harness it not to destroy DNA molecules but, in fact, to rewrite them.”
Nowadays, Dr. Doudna’s team of researchers at UC Berkeley is working to refine CRISPR/Cas9 as a gene editing tool. By investigating the Cas9 enzyme’s 3D structure, for example, her team hopes to better understand how Cas9 works and even make changes to it to improve its accuracy and avoid altering DNA at undesired locations. At the same time, her group is scouring the microbial world for new CRISPR/Cas systems. This effort has uncovered three new CRISPR/Cas systems, including two that rely on never-before-seen proteins Dr. Doudna has dubbed CasX and CasY. Finally, Dr. Kyle Watters, a postdoctoral researcher in Dr. Doudna’s lab, is using a computer algorithm he designed to identify sequences in bacterial genomes that code for Cas-inhibiting proteins. These molecules could help researchers better control CRISPR/Cas-based gene editing by preventing the Cas protein from targeting certain DNA sequences and by permitting scientists to shut down Cas proteins after they have finished making the desired genetic changes.
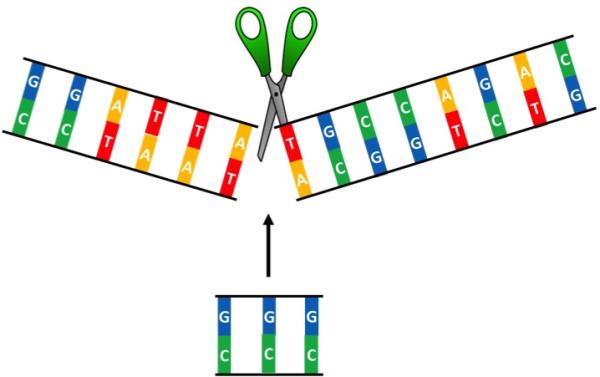
CRISPR/Cas9 revolutionized gene editing due to its ability to cut double-stranded DNA at specific locations, allowing scientists to insert or remove segments of DNA as they see fit.
Of course, even in the midst of Dr. Doudna’s many basic research studies on how CRISPR/Cas systems function, she has not lost sight of the equally important work of using the technology to improve human health. In this arena, her group is currently focused on using CRISPR/Cas9 to edit genes in the brain in order to treat neurological diseases. Experiments being led in her lab by postdoctoral researcher Dr. Brett Staahl aim to chemically modify the Cas9 protein to allow it to enter neurons once it has crossed the blood-brain barrier. Other scientists outside Dr. Doudna’s lab are hard at work deploying CRISPR/Cas9 to edit animal genes so that their organs might be safely transplanted into human patients who need a new heart, kidney, or liver. In the nearer term, because of CRISPR/Cas9’s ability to locate and interact with specific DNA and RNA sequences, the system could be used to diagnose cancers and infections by detecting genetic material from viruses, bacteria, and tumors. A collaboration between Dr. Doudna and Dr. Joel M. Palefsky at the University of California, San Francisco, for example, is attempting to use CRISPR/Cas9 to diagnose human papilloma virus (HPV), which can cause cervical cancer.
“Applications are going to require very careful thought, and we need to do things like developing better ways of delivering these molecules into cells and tissues and also [of controlling] them once they get there,” Dr. Doudna said. “The fundamental biology is going to continue to drive this technology forward. I think that’s what we’ve always seen: that the fundamental science and sort of curiosity-driven experimentation goes hand-in-hand with the technology development.”
With all the tantalizing applications scientists have dreamed up for CRISPR/Cas systems, it can be easy to forget the profound ethical questions conjured up by the possibility of manipulating the human genome. Dr. Doudna has also done important work on these issues, such as her contributions at the first International Summit on Human Gene Editing, which took place in Washington. D.C. in 2015. The meeting produced a consensus statement detailing a path forward for human gene editing, with a particular emphasis on altering genes in reproductive cells like sperm and eggs in which the changes would be passed on to future generations. Dr. Doudna is looking forward to the follow-up to that meeting, which is planned for this November in Hong Kong.
“The technology is moving ahead at a breakneck pace,” Dr. Doudna said, “and so that really does require us to think very carefully about how we’re going to deploy this technology going forward.”
Subscribe to our weekly newsletter to stay up-to-date on the latest breakthroughs in the NIH Intramural Research Program.
Related Blog Posts
- IRP’s Thomas Kunkel Elected to National Academy of Sciences
- IRP’s Eugene Koonin Elected to National Academy of Medicine
- Award Honors Promising Female Scientists
- Global Scientists Come Together at the National Institutes of Health
- RNA-Targeting Therapeutic Restores Protein Absent in Spinal Muscular Atrophy
This page was last updated on Monday, January 29, 2024
