Pushing the Frontiers of Imaging
Advanced Imaging and Microscopy Facility Opens in Building 13
The walls are freshly painted, new shelves have been installed, and the microscopes are being moved in. The sparkling new Advanced Imaging and Microscopy (AIM) facility in Building 13 on the NIH Bethesda campus is open for business. It’s a trans-NIH core facility that houses, operates, disseminates, and improves noncommercial, prototype optical-imaging systems.
In fact, AIM has been operating for more than three years out of temporary quarters in Hari Shroff’s lab (in Building 13) at the National Institute of Biomedical Imaging and Bioengineering (NIBIB).

Hari Shroff.
The seeds of the AIM group were planted almost five years ago during a lunchtime conversation among four intramural imaging scientists: Shroff, Justin Taraska and Keir Neuman (National Heart, Lung, and Blood Institute), and Dan Larson (National Cancer Institute). They wanted to push the boundaries of imaging technology to enhance intramural research projects. Wouldn’t it be great, they thought, to have a central core facility—available to all intramural researchers—that could develop cutting-edge microscope technology and new image-processing algorithms?
It took a couple of years for the proposal to be approved and intramural funds secured. Shroff, the managing director of the core facility, has overseen AIM from its inception. Taraska, Neumann, and Larson are on the AIM steering committee along with representatives from other NIH institutes.
The AIM Microscopes
AIM’s goal is to be a constantly evolving facility, with the microscopes updated, modified, and replaced with newer technology on a continuing basis.
Here’s a list of what AIM has on hand now:
Dual-view Inverted Selective Plane Illumination Microscope (diSPIM)
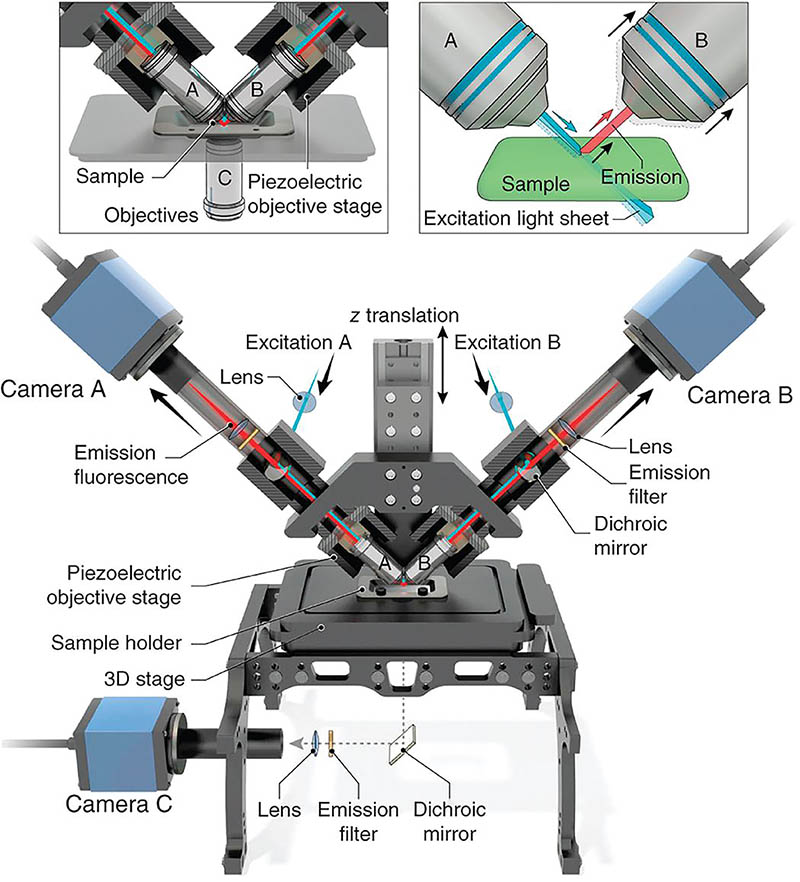
CREDIT: AIM FACILITY, NIBIB
In 2013, Shroff developed the diSPIM, which is a dual light-sheet, fluorescence microscope that acquires two perpendicular views of a sample. It can quickly image live cellular processes—such as viruses moving quickly, cancer cells migrating, and neurons interacting—at high resolution and in 3-D without causing extensive light damage. The diSPIM has two cameras (traditional microscopes have only one) that acquire two views of a sample. These views are then fused together in a computer. The technology has been patented and licensed by NIH.
Instant Structured Illumination Microscope (iSIM)/Total Internal-reflection Fluorescence SIM (TIRF-SIM)
The iSIM is another fluorescence microscope developed in the Shroff lab. The iSIM collects images rapidly, with twice the resolution of a confocal microscope and in three dimensions. A 2018 modification to the microscope also offers TIRF capability for very-high-contrast imaging near the cell surface. This technology has also been patented and licensed by NIH.
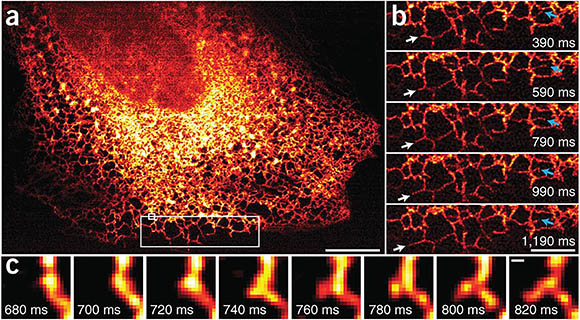
CREDIT: AIM FACILITY, NIBIB
Images taken by the Instant SIM reveals endoplasmic reticulum (ER) dynamics within human lung fibroblasts. (a) First image in series of 200 time points; (b) Higher-magnification view of the large white rectangle in a. White arrows mark growth of an ER tubule; blue arrows indicate remodeling of an ER tubule; (c) Higher-magnification view of the small white rectangle in a, indicating formation of a new tubule.
Interferometric Photoactivated Localization Microscope (IPALM)
Donated by Taraska, the IPALM combines photoactivated localization microscopy (a super-resolution technique) with interferometry (a technique that superimposes electromagnetic waves and uses the interaction between them to provide very sensitive distance measurements), providing less than 20-nanometer resolution in all three dimensions. “Incidentally,” said Shroff, “the same technique [interferometry] is used to detect gravitational waves.”
High-throughput Single-molecule Imaging Microscope
Donated by Larson, this microscope was designed to image tens of thousands of cells and can identify and count nuclei, cell bodies, and molecules.
Single-molecule TIRF/Inclined-illumination Microscope
This microscope can be used to examine single-molecule interactions or biomolecules in live cells.
Collaborations
The AIM group did not wait for their facility to be completed before diving into collaborations. Shroff and Harshad Vishwasrao, the first hire (in 2016), began to give energetic, exciting seminars around campus that prompted a slew of intramural scientists to contact them about potential collaborative projects. A sampling of these projects is described here.
Imaging mouse eggs
Jurrien Dean at the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) became one of these collaborators. His lab studies mouse sperm-egg interactions and the postfertilization block to polyspermy. Only one sperm is needed to fertilize an egg; two or more sperm are lethal to the development of an embryo. The egg, however, can protect itself. After fertilization, the egg’s peripheral cortical granules (CGs) release ovastacin, an enzyme that modifies the egg’s outside layer in a way that prevents additional sperm from binding. Edgar-John Vogt, a postdoc in Dean’s group, used ovastacin, tagged with a fluorescent probe, to learn more about CG biology. But he and Dean needed super-resolution imaging to properly study the CGs. NIDDK’s own confocal microscopes weren’t enough. Luckily, Dean had heard Shroff give a presentation and therefore knew about AIM’s sophisticated microscopes. He also was impressed by Shroff’s enthusiasm and interest in cutting-edge projects, and so a collaboration was born.
AIM’s advanced imaging—TIRF, iSIM, and live instant TIRF-SIM—allowed the researchers to see CGs at a single-granule resolution. These images documented for the first time that the myosin IIA protein was involved in transporting CGs to the egg’s plasma membrane and actin proteins were cleared to facilitate CG fusion with the membrane. The researchers also demonstrated that a large protein called maternal antigen that embryos require (MATER) helps to anchor the CGs in the cortex of the egg. In the absence of MATER, the CGs are present throughout the egg instead of being near the membrane where they can keep multiple sperm from fertilizing the egg.
Vogt especially appreciates that the AIM group taught him how to use their microscopes, helped him troubleshoot problems, and let him independently collect data.
Although Dean’s project imaging the CGs has been completed, he said that high-resolution imaging of these delicate biological structures and the expertise and technology offered by the AIM facility will continue to be important for his lab’s research.
Imaging the retina
At the National Eye Institute, Anand Swaroop heard of the AIM group and invited Vishwasrao to give a talk to his lab. Swaroop is trying to understand the molecular and cellular pathways underlying retinal development, aging, and disease, and developing new treatments for blinding retinal diseases. His lab intends to collaborate with AIM on two or three new projects. One will examine details of the human retina made in a dish from patients’ cells. In another project, Swaroop’s group is studying the retina in several vertebrate species to determine how rods (responsible for vision at low light levels) and cones (active at high light levels and responsible for color vision) develop, how color vision evolved, and how the cone-rich retinal fovea that provides sharp central vision develops. Swaroop’s work has tied the development of rod photoreceptors to the evolution of early mammals from nocturnal creatures to diurnal ones. Discovering how human vision evolved will also advance the study of retinal and macular degeneration.
To further their work, the researchers need histological data from many species at super-high resolution. Noor White, a postdoc in Swaroop’s lab, has started using the AIM diSPIM, with Vishwasrao’s help, to collect images of the retina from birds as part of her research on the evolution of night vision and development of the fovea.
The AIM group is “an amazing facility with very enthusiastic people who are interested in challenging themselves with novel projects,” said Swaroop.
Imaging of bacterial spore coats
AIM Co-director of Operations Jiji Chen, who specializes in single-molecule imaging techniques, collaborated with Shroff and Kumaran Ramamurthi’s group at the National Cancer Institute to determine how bacterial spore coats form. The process is a model for understanding the dynamics of structural proteins as they organize in the correct place and time during morphogenesis. In a recent study, the researchers described how the bacterial spore coat, which consists of dozens of proteins, assembles in an orchestrated fashion. Images collected on the AIM single-molecule TIRF microscope showed how these coat proteins recognize and bind to membranes with a specific curvature. (Proc Natl Acad Sci U S A 116:21789–21799, 2019; DOI:10.1073/pnas.1907397116).
Lending technical expertise to build specialized microscopes
AIM researchers are engaged in purely technical collaborations, too, such as one with Ted Usdin, the director of the Systems Neuroscience Imaging Resource in the National Institute of Mental Health (NIMH). He wants to image large volumes of brain tissue to examine the connections, distributions, and phenotypes of neurons. His group, and others at NIMH, would like to image the entire brains of mice and rats—and potentially even large primate brains—at high resolution.
NIMH has funded the development of a specialized, cleared-tissue diSPIM—to be housed in and operated by AIM—that can rapidly visualize entire mammalian brains. NIMH investigators are eager to use this instrument because in contrast to other light-sheet instruments there is no theoretical limit to the lateral dimensions of the tissue that can be imaged. The instrument will be able to image material that is up to 50 x 75 millimeters (mm) in length and width and 2 mm thick. A cutting device called a vibratome, mounted onto the microscope stage, will successively remove 1–2 mm of tissue at a time after each section is imaged. An entire brain can be imaged this way. Vishwasrao will be building and maintaining the microscope as well as making design changes when necessary. The microscope is expected to be ready in the next few months. The images will allow NIMH scientists to examine the connections that neurons make among brain regions and identify particular cells involved in these connections.
“The big picture is correlating brain structure and activity with behavior,” said Usdin. “Being able to look at the neuronal connections will facilitate those experiments.”
Neural sample imaging (multi-institute consortium)
CREDIT: SUSAN CHACKO, CIT
From left: Mark Hoon (not shown) and Nick Ryba at the National Institute of Dental and Craniofacial Research, Claire Le Pichon at the National Institute of Child Health and Human Development, and Alexander Chesler at the National Center for Complementary and Integrative Health are using diSPIM to combine imaging of neural samples with gene-expression data.
Mark Hoon and Nick Ryba at the National Institute of Dental and Craniofacial Research (NIDCR), Claire Le Pichon at the National Institute of Child Health and Human Development, and Alexander Chesler at the National Center for Complementary and Integrative Health (NCCIH) wanted to combine imaging of neural samples with gene-expression data. Then they heard about the diSPIM technology. Discussions with the AIM group followed, and in 2019, the consortium was awarded Director’s Challenge Award funds (to supplement funds from NIDCR and NCCIH) to develop a very fast in situ microscope to track a neural sample’s response to stimuli. The new instrument will be a technical extrapolation of an existing AIM diSPIM, capturing images rapidly at a rate of 10 milliseconds per image with a large field of view, but with twice the resolution. The samples will be stained with fluorophores that bind to specific RNA sequences and emit light at multiple wavelengths, and the microscope will have the ability to fully resolve many fluorophores. The AIM group provided the expertise for the development of the microscope and will collaborate on troubleshooting and further development, but the device is housed and managed by the other members of the consortium.
Already largely built as of December 2019, the new microscope will initially be used to image specific sensory neurons in the trigeminal ganglion that respond to stimuli such as touch, temperature, and a range of chemicals. The consortium researchers plan to combine the new imaging data with their ongoing single-cell gene-expression experiments with long-term goal of understanding how pain signals are processed. Their investigations will include exploring sensory deficits, the response of neurons to injury, chronic neuropathic pain, and disease states or genetic anomalies that alter the sensation of touch and temperature.
Challenges
With new technological developments come new challenges. The problem of data processing was noted by both Vishwasrao and Usdin. Although collecting a dataset with the high-powered microscopes can take minutes, processing the data can take weeks. Some of the processing will be done on Biowulf, NIH’s biomedical supercomputer, but transferring, storing, processing, and archiving multiterabyte datasets can all be bottlenecks.
Luckily, the AIM group is also developing image-analysis software that will speed up processing. A recent addition to the AIM staff is Jiamin Liu, who will be using artificial intelligence and deep-learning techniques to enhance image quality by deblurring and denoising them (noisy images are imperfect because of random digital or electronic noise caused by the camera sensor); and to improve image analysis.
How AIM is funded
At present, the AIM facility operates on a partial cost-recovery model where 25% of the costs are covered by the collaborators. Some institutes pay out of a central fund while others charge the individual collaborating labs, adding a level of complexity to the charging procedures. Over time, the AIM will be required to recover a larger fraction of its costs, but Shroff is concerned that higher charges will discourage exciting new projects, especially those that entail risk.
Long history
The AIM core facility is but one of the several new ventures in NIH’s long history of pioneering imaging techniques. In the 1960s, for example, the real-time picture processor was among the first computer-hardware systems designed for imaging.
“The beauty of the NIH is having these pockets of expertise in areas where normally our groups would not interact,” said Dean, “but when you have the need, you can go and take advantage of it.”
For more information about AIM and how it may be able to help you, visit its website at https://www.nibib.nih.gov/labs-at-nibib/advanced-imaging-and-microscopy-aim-resource. AIM is but one of several core imaging facilities. Read Michael Gottesman’s essay in this issue of the NIH Catalyst for descriptions of others.
This page was last updated on Tuesday, March 29, 2022
