Wonder Drugs and Super Bugs
The Rise of Antibiotics…and Antibiotic Resistance
Ever since 1928 when penicillin mold was discovered to secrete an antibacterial substance, doctors have been developing antibiotics to fight bacterial infections. Now the bacteria are fighting back. But NIH scientists are seeking ways to thwart antibiotic-resistant bacteria, developing new antibiotics that work differently than the older ones, and even trying to understand antibiotic resistance from a historical perspective.
“I don’t think we can fully appreciate our present options with respect to global antibiotic development, usage, and resistance without a deeper understanding of the historical forces that have brought us to this point,” Harvard medical historian Scott Podolsky said in an interview with the National Library of Medicine’s (NLM’s) Circulating Now blog. In November, Podolsky, who is the director of Harvard’s Center for the History of Medicine (Boston), delivered an NLM History-of-Medicine lecture—“Antibiotic Pasts and Futures: Seven Decades of Reform and Resistance”—based on his research at NLM for a new book that has just been published.
In his talk, Podolsky provided a whirlwind tour of the history of antibiotics from the 1940s on, exploring the evolving relationships among industry, academia, medicine, regulators, and the public. One of the key themes was concern over antibiotic resistance (AR). AR isn’t a new problem—we have known about the ability of bacteria to evolve in response to selective pressure from antibiotics since the beginning. However, an inherent lack of stewardship—namely, the overuse and misuse of antibiotics both in the human population and in animals used for food—has put the AR process in overdrive.
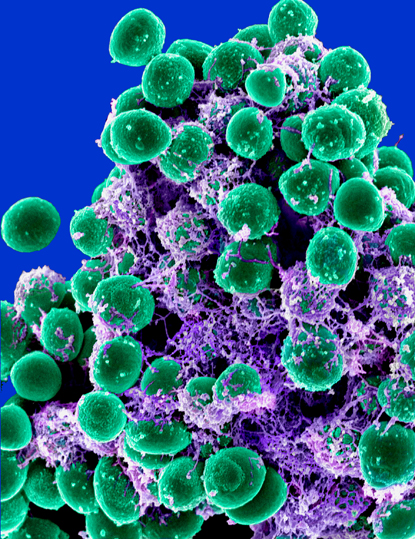
ELECTRON MICROSCOPY UNIT, ROCKY MOUNTAIN LABS, NIAID
Michael Otto (NIAID) studies the mechanisms of pathogenicity in Staphylococci, including the formation of multicellular bacterial agglomerations called biofilms. Antibiotics often cannot penetrate these dense, sticky matrices, and even if they do, the metabolism of these cells is such that they are not susceptible to the drugs. Shown: Scanning electron microscopy of a Staphylococci biofilm (balls) embedded in an exopolysaccharide matrix.
In the United States alone, antibiotic-resistant bacteria are estimated to cause at least two million infections a year and 23,000 deaths; many more people die from other conditions complicated by an antibiotic-resistant infection (http://1.usa.gov/1n5K4VF). AR has become a national security and public health issue, which is all the more worrying because at the same time the antibiotic-development pipeline needed to replenish our arsenal has effectively dried up, according to Amy Patterson, former NIH associate director for Biosecurity and Biosafety Policy.
“Combatting antibacterial resistance is now a national and indeed an international global public-health priority,” said Patterson, at a July 2014 NIH workshop on the development of new antibacterial products (http://videocast.nih.gov/launch.asp?18549). “Addressing the issue will require efforts in a variety of areas such as antibiotic stewardship, disease surveillance, [and] basic and applied research as well as ultimately new diagnostics and therapeutics.”
At the same meeting, NIH Director Francis Collins emphasized that “AR is a high priority for this administration.” In fact, on September 18, 2014, President Obama issued an Executive Order releasing the National Strategy for Combating Antibiotic-Resistant Bacteria (CARB) and directing the government to take action to combat the rise in AR (http://1.usa.gov/ZrXYwP)
The NIH is taking a multifaceted approach to addressing this challenge. To galvanize the research and development community, the NIH and the Food and Drug Administration have jointly sponsored workshops exploring issues related to antibacterial product development, including public-private partnerships and the streamlining of clinical trials and regulatory pathways. NIH will also offer a $20 million prize to facilitate the development of a rapid point-of-care diagnostic test to identify highly resistant bacterial infections (http://1.usa.gov/1wMF0yM). This initiative will aid both the surveillance and more judicious use of antibiotics.
Several intramural PIs in different institutes work on AR, some with grants from the Director’s Challenge Innovation Award Program, which in 2013–2014 supported 10 innovative intramural projects on AR (http://sigs.nih.gov/challenge/Pages/FundedProjects.aspx). Julie Segre, chief of the Translational and Functional Genomics Branch at the National Human Genome Research Institute (NHGRI), received one of these awards for her work studying the microbiome of Clinical Center (CC) patients who are at risk of infection with multidrug-resistant bacteria.
“Hospital infections are one of the looming crises facing the delivery of health care,” she said. With the paucity of new antibiotics in the pipeline, hospital infection control is crucial in the fight against multidrug-resistant organisms such as carbapenem-resistant Klebsiella pneumoniae (KPC).
Segre uses genomic sequencing to study modes of transmission in microorganisms such as bacteria and has leveraged these techniques to tackle AR. In 2011, she collaborated with CC epidemiologists Tara Palmore and David Henderson and NHGRI bioinformatics specialist Evan Snitkin to lead the NIH’s response to the spread of KPC, which resulted in the deaths of several patients at the CC. The team used whole-genome sequencing of the bacterium with epidemiological analysis to track the spread of KPC and to determine why it progressed in spite of the early implementation of infection-control procedures.
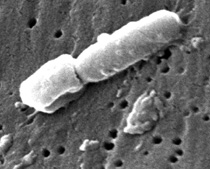
JANICE CARR, CDC
This scanning electron micrograph reveals some of the ultrastructural morphologic features of a Klebsiella pneumoniae bacterium that Julie Segre and others are studying.
Their findings provided evidence for unexpected transmission routes (for example, some patients who did not display symptoms were actually carriers) and were used to drive a change in infection-control procedures (Sci Transl Med 4:148ra116, 2012).
“Samples used in surveillance for AR bacteria are now collected on admission to the hospital, twice weekly in the intensive-care unit, and monthly from all inpatients,” said Segre. Ongoing surveillance by her lab shows that there have not been any transmissions of KPC for two years in the CC.
In recognition of their efforts, Segre and other members of the CC response team received Samuel J. Heyman Service to America Medals (Sammies) for “stop[ping] the spread of a deadly hospital-acquired infection through the first-ever use of genome sequencing to identify the source and trace the transmission of antibiotic-resistant bacteria, creating a groundbreaking model for the health-care industry.”
National Institute of Diabetes and Kidney Diseases (NIDDK) Senior Investigator Carole Bewley also refocused her research after the KPC outbreak. Bewley can be thought of a drug hunter of sorts, extracting novel molecules from natural sources and testing their potential as antibiotics.

PAT COLIN, CORAL REEF RESEARCH FOUNDATION, PALAU
Carole Bewley collecting samples at Blue Hole Cave in Palau. On this trip, her lab collected the sponges that have yielded the novel antibiotics that Bewley’s group is studying.
“We are trying to discover new antibiotics that have either different mechanisms of action or different chemical scaffolds (structures) so that they will hit a target on a bacterium that is resistant to known antibiotics,” she said. For example, Bewley’s lab discovered a rare species of marine algae that produces a substance that kills all drug-resistant forms of gram-positive bacteria such as methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococcus faecium.
Bewley has been collaborating with CC Microbiology Chief Karen Frank (who also has a Director’s Challenge Innovation Award) to test the effectiveness of novel antibiotic isolates against antibiotic-resistant bacteria such as KPC. Bewley’s lab has identified several compounds that are effective because they have a novel target on the bacterium; her lab is now doing whole-genome sequencing to try to work out what the target actually is.
Although research into AR is very much a trans-NIH venture, the National Institute of Allergy and Infectious Diseases (NIAID) is responsible for the lion’s share of NIH’s basic, translational, and clinical research in AR. NIAID considers its program a “race to outsmart bacteria by working around the mechanisms that cause resistance,” according to the AR Strategic Plan. (http://www.niaid.nih.gov/topics/antimicrobialResistance/Documents/ARstrategicplan2014.pdf). Several intramural NIAID PIs conduct AR research, including molecular microbiologist Michael Otto, chief of the Pathogen Molecular Genetics Section in NIAID’s Laboratory of Human Bacterial Pathogenesis.
Otto studies the mechanisms of pathogenicity in Staphylococci, including the formation of multicellular bacterial agglomerations called biofilms. Antibiotics often cannot penetrate these dense, sticky matrices, and even if they do, the metabolism of these cells is such that they are not susceptible to the drugs. “AR in biofilms is categorically different [from] other mechanisms of resistance [such as] a bacterial enzyme that degrades an antibiotic or a pump that pumps antibiotics out of the cell,” Otto explained. “Biofilms confer resistance in a nonspecific way, therefore resisting all antibiotics.” Understanding how biofilms work may help scientists develop drugs or vaccines that interfere with their immune-evasion mechanisms, providing alternatives to antibiotics.
In a related line of research, Otto has been collaborating with another NIAID PI, Yasmine Belkaid, chief of the Mucosal Immunology Section in the Laboratory of Parasitic Diseases. They are investigating the interactions between the “bad bacteria” such as Staphylococcus aureus and the “good bacteria” that inhabit our bodies to see whether their benefits might extend to combatting Staphylococci. Belkaid is also collaborating with Frank and Segre to develop a mouse model of KPC infection.

In 2013, the team of NIHers that worked on an antibiotic-resistant form of the Klebsiella bacteria received the Samuel J. Heyman Service to America Medals (Sammies) for “stop[ping] the spread of a deadly hospital-acquired infection through the first-ever use of genome sequencing.” From left: Julie Segre, Evan Snitkin, Tara Palmore, ABC News political correspondent Cokie Roberts, and David Henderson.
AR is clearly an urgent global issue, perhaps as big as the AIDS problem was in the 1980s, said Otto.
These sentiments were echoed by Podolsky. AR is a “ticking time bomb,” he said. “There has been attention [paid] to AR for the past 30 years—what we need now is more than just attention…we need to galvanize action!”
The good news is that we are making progress on this front. “There has been an upswing in interest, so the future is bright,” said Bewley. “I think that what needs to happen is cross-fertilization across research disciplines.”
As Collins eloquently stated in the antibacterial products workshop, “I envision a future where we stay one, two, or even three steps ahead of AR by working together to develop innovative solutions that we haven’t even dreamed of yet.”
To read an interview with Scott Podolsky on NLM’s Circulating Now blog, go to http://1.usa.gov/1BjZiRI [http://circulatingnow.nlm.nih.gov/2014/11/05/antibiotic-pasts-and-futures]. His new book is The Antibiotic Era: Reform, Resistance, and the Pursuit of a Rational Therapeutics (Baltimore: The Johns Hopkins University Press, 2014).
This page was last updated on Tuesday, April 26, 2022
