No Footprints?
NIH Scientists Discover Possible Flaw in an ENCODE Trusted Technique
NIH researchers have found a flaw in an important tool that is supposed to identify certain functional elements of the human genome. The finding, that some proteins bind too briefly with DNA to leave “footprints,” may prompt a rethinking of how best to map regulatory regions in DNA.
Although the human genome was fully sequenced in 2003, the function of most of its three billion base pairs is unknown. Only about one percent of the pairs is in protein-coding regions; the rest of the genome contains other functional (but non-protein-coding) elements that turn genes on or off, delineate chromatin structure, or sequences that produce regulatory RNA molecules.
The daunting task of identifying all the protein-coding and noncoding areas is akin to assigning street- and household-level information to satellite images of towns and cities around the globe. To meet the challenge, the National Human Genome Research Institute organized a consortium of 32 international genomics laboratories to collaboratively build an ENCyclopedia of DNA Elements, or ENCODE, which would systematically map the precise location of all protein-coding and non-protein-coding functional elements within the genome.
CREDIT: M. SUNG AND G. HAGER, NCI
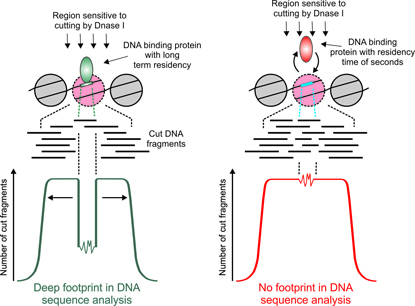
Among the many ENCODE methods used to identify DNA elements—both protein-coding and non-protein-coding—is genomic footprinting. This method involves cutting chromatin with the enzyme DNase I and mapping accessible regions by sequencing. A bound protein will protect a short sequence from DNase I and leave a “footprint” in the computational analysis. NIH scientists led by Gordon Hager, however, have discovered that only proteins that bind DNA for longer periods of time leave detectable footprints (left). Some proteins, such as the many transcription factors that bind to DNA for shorter amounts of time, do not leave footprints (right). Consequently, genomic footprinting cannot be used to predict DNA-binding patterns for numerous transcription factors.
Ultimately, ENCODE is to help scientists understand how genomic information is choreographed to create a complex organism and how that choreography can go awry in disease.
Among the many methods used to identify the DNA elements is genomic footprinting, also called digital genomic footprinting. This method is an extension of DNase-seq, which involves cutting chromatin with the enzyme DNase I and mapping accessible regions by sequencing. Within these “open chromatin” regions a bound protein will protect a short sequence from the DNase I and leave a “footprint” in the computational analysis. One set of ENCODE elements, published in 2012, relied on the footprinting tool to create an extensive map of locations where transcription factors bind to DNA to control the reading of the genomic information at protein-coding or RNA-coding sites.
Gordon Hager in the National Cancer Institute and his colleagues were surprised to discover that footprinting analysis failed to detect binding sites for proteins that only briefly bind to DNA. Hager studies the action of steroid receptors, including the glucocorticoid receptor (GR) and the estrogen receptor (ER), which act as transcription factors when bound by specific hormones. His work, using diverse experimental methods including biochemistry and single-molecule imaging studies, shows that these receptors interact surprisingly transiently with their DNA targets, for roughly 10 seconds. And they don’t leave footprints!
To understand why, staff scientists Myong-Hee Sung and Songjoon Baek in Hager’s group developed a sensitive footprint-detection algorithm called DNase2TF. Using the ENCODE data as well as other published DNase footprinting data, they blind tested the software for footprint detection and evaluated their predictions against independently confirmed transcription-factor binding sites. Their software was able to predict transcription-factor sites more effectively than all the available footprint-detection algorithms, but it still could not detect footprints at a large number of confirmed transcription-factor binding sites, including the GR-binding elements.
With postdoctoral fellow Michael Guertin, they confirmed that many dynamic transcription factors such as GR, ER, and serum response factor, a transcription factor involved in cell growth and differentiation, also bind DNA without an associated footprint. Only transcription factors with longer DNA residency times generate footprints. The well-studied transcription-factor CTCF (with a DNA residency time of about five minutes) leaves deep footprints, whereas other factors that bind DNA longer than GR, but more briefly than CTCF, leave shallower ones.
Hager’s group is currently strengthening this correlation between DNA association time and footprint depth by analyzing the footprints and binding dynamics of a range of transcription factors.
This work demonstrates that footprinting is “not yet a mature methodology,” said Sung.
The current next-generation sequencing produces unprecedented amounts of genomic data from ENCODE (and elsewhere). Mining meaningful information and patterns in the data requires not only sophisticated software and computational tools, but also an integration with knowledge gleaned from other disciplines. Hager’s collaboration with imaging laboratories and their integration of data from a wide range of experimental systems have been vital in capturing the in vivo behavior of transcription factors.
To fully understand the complexities of the human genome and protein dynamics, more collaborative efforts among scientists in different disciplines are needed. “We can’t work alone in silos,” said Hager. “The genomics people need to work with the biochemists and with the single-molecule imaging experts.”
Hager’s study can be read in Molecular Cell (Mol Cell 56:275–285, 2014).
The ENCODE footprinting study can be found in Nature (Nature 489:83–90, 2012).
DNase2TF is available at http://sourceforge.net/projects/dnase2tfr/.
This page was last updated on Tuesday, April 26, 2022
