The Human Microbiome Project
… And the Intramural Connection
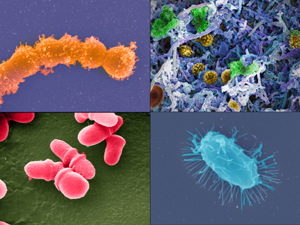
CREDIT: Jonathan Bailey, NHGRI
The Human Microbiome Project, which was launched by NIH in 2007, provided the first glimpse of the microbial diversity of healthy humans and is exploring the possible relationships between particular human diseases and the microbiome. (Clockwise from top left): Streptococcus (Credit: Tom Schmidt); microbial biofilm of mixed species, from human body (Credit: A. Earl, Broad Institute/MIT); Bacillus (Credit: Tom Schmidt); Malassezia lopophilis (Credit: J.H. Carr, CDC).
There are 10 times as many bacteria, viruses, fungi, and protozoa–collectively known as the microbiome–living on and inside the human body as there are human cells. Although scientists have been aware of the microbiome for more than 30 years, they knew little about its diversity and role in human health and disease. Researchers tended to focus on disease-causing bacteria, and only 10 to 20 percent of these bacteria can be cultured in the laboratory.
In 2005, at an international meeting in Paris, scientists proposed using state-of-the-art genomic sequencing techniques to catalogue all the bacteria living on and inside the human body. They also discussed forming a consortium to catalogue the intestinal microbiome and its role in human health and disease.
Phase One of the HMP focuses on a survey of the microbiome in five areas of the body—the digestive tract, mouth, skin, nasal cavity, and vagina—and consists of two kinds of cohort studies: the Healthy Cohort Study and a collection of Demonstration Projects. The second phase of HMP which started on September 8, 2013, focuses on a survey of the biological properties of the microbiome, such as gene expression profiles, proteins, and metabolites, produced by the microbiome under specific conditions such as in preterm birth or in diabetic patients.
The goal for the Phase One Healthy Cohort Study was to create a reference microbiome that represents the normal or standard collection of bacteria living on and inside healthy American adults. This study has provided the first glimpse of the microbial diversity of healthy humans; the major findings are described at the end of this article in an interview with Lita Proctor and online at http://www.hmpdacc.org.
The goal for the Demonstration Projects was to explore the possible relationships between particular human diseases and the microbiome. Researchers collected and sequenced microbiome samples from diseased volunteers and compared them with the reference microbiomes from normal, healthy volunteers. They found that the microbiome from individuals with a disease—such as Crohn’s Disease, eczema, or esophageal adenocarcinoma—is significantly different from those of healthy individuals. Scientists are still trying to determine whether the microbiome differences are a cause or effect of disease.
Intramural Microbiome Research
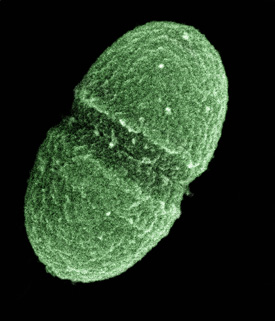
U.S. Department of Agriculture
The bacterium, Enterococcus faecalis, which lives in the human gut, is just one type of microbe that is being studied as part of NIH’s Human Microbiome Project.
What follows are descriptions of three Intramural Research Program activities in the HMP.
Julie Segre, senior investigator, NHGRI, and Heidi Kong, investigator, NCI-CCR: The microbiome of the skin
Julie Segre and Heidi Kong have played a large role in helping us to understand the microbiome of the skin. Our skin is a diverse ecosystem analogous to Earth’s various ecosystems (oily versus dry, for example). What Segre and colleagues have found is that the moist areas of the body have the greatest number of bacteria but the least diversity, while the dry areas of the skin have the fewest bacteria but the most diversity. Segre’s laboratory studies atopic dermatitis (eczema) in young children, and she hopes understanding the microbiome of the skin will lead to improved treatments. Atopic dermatitis is characterized by asymptomatic periods punctuated by periods of severe skin inflammation.
Segre and Kong found that the microbial population changes during dermatitis flares, and they are exploring whether the microbial diversity can be used to predict when dermatitis flare-ups will occur. In addition, there are multiple treatments for atopic dermatitis and not all children respond similarly. Because there is no adequate diagnostic to predict which treatment will work best, Segre and Kong are investigating whether the microbial diversity of atopic dermatitis can be used to identify the most effective treatment.
Jason Brenchley, senior investigator, NIAID: Microbiome of the intestine
It may not seem intuitive, but the gut microbiome plays an important role in human immunodeficiency virus (HIV) infection. Jason Brenchley has played an integral role in understanding this complex interaction. Although antiretroviral therapy has proven successful in controlling the replication of HIV in the blood and in reducing the incidence of acquired immunodeficiency syndrome (AIDS), the life expectancy of infected, but treated, individuals is shorter than that of healthy individuals. Recent data have shown that the cause of death in HIV patients is strongly associated with inflammation and microbial translocation from the gastrointestinal (GI) tract lumen. During the acute phase of HIV infection there is a significant depletion of GI-tract-resident CD4+ T cells, apoptosis of gut epithelial cells, and a shift away from the normal immune-regulating cells. These changes make the gut epithelium susceptible to microbial translocation into the body, causing chronic low-grade systemic inflammation that over time can result in cardiovascular disease. Using an animal model of HIV, Brenchley has found that bacteria that translocate across the gut epithelium are not representative of the gut microbiota and that bacteria of the phylum Proteobacteria (including invasive strains such as brevundimonas and diaphorobacter) tended to translocate preferentially.
These studies led to a proposal for using probiotics (healthful bacteria) to increase the percentage of beneficial, noninflammatory microbes in the gut, thus reducing the percentage of invasive proteobacteria. The probiotics produce factors that act on the gut epithelium to restore its structural integrity, reduce inflammation, and prevent microbial translocation. Ongoing studies in Brenchley’s laboratory aim to understand how HIV infection alters the gut microbial composition and metabolism.
Yasmine Belkaid, senior investigator, NIAID: Microbiome of infection barrier sites (skin and gut)
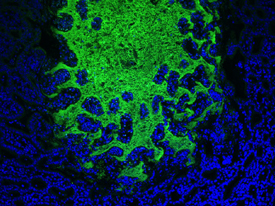
YASMINE BELKAID, NIAID
Microbiota in the gut (green) and the epithelial cells (blue).
Harmful bacteria and viruses gain entry to the body by traversing its protective barriers such as the skin, mucosal epithelium of the nasal passages, and the gut epithelium. Until recently it was thought that these protective barriers (especially the skin) functioned independently of the resident or commensal microbiota known to reside on them. We now know that the commensal microbiota of the barrier sites are integral in keeping harmful bacteria from entering our body.
Yasmine Belkaid’s laboratory, in collaboration with Julie Segre (NHGRI) and Heidi Kong (NCI-CCR) was instrumental in demonstrating the importance of the skin microbiome as a barrier against infection. She compared the response of two groups of mice to infection. The first group was housed normally and were host to the normal array of commensal microbiota; the second group was housed from birth in a highly controlled environment free of germs, including commensal microbiota. When the two groups were exposed to the pathogenic bacteria Leishmania major (which is transmitted by sandflies and causes cutaneous leishmaniasis), the normal group mounted a normal immune response whereas the sterile-environment group failed to mount such a response. Additional experiments demonstrated that the skin’s commensal microbiota are essential in priming the skin’s immune system to respond to noncommensal pathogenic microbiota.
Major Findings of the HMP
An edited interview with Lita Proctor, HMP program officer, NHGRI
What has been the most unexpected finding from the HMP?
The sheer magnitude of the genetic potential present in the thousands of microbes living on and inside the human body. Humans are hosts to 10,000 different bacterial species, and each individual carries around 1,000 different species. The global pool of unique microbial genes associated with the 10,000 microbial species in healthy individuals is estimated to be around eight million. In stark contrast, humans only have 23,000 genes. Since the human microbiome field is still in its infancy, researchers know very little about the function of those bacterial genes.
What benefits can patients expect to see from data generated from the HMP?
Indirect benefits of the HMP are already being realized. Fecal microbiota transplantation, the process of transplanting fecal bacteria from a healthy individual into a recipient individual, is frequently used to treat the symptoms of Clostridium difficile infection (such as antibiotic-associated diarrhea). Doctors are now using the sequencing technology and analysis developed for the HMP to perform sequenced-based analysis of donor and recipient stool to predict how individuals will respond to more-tailored treatments. The long-term benefits of the HMP are predicted to be wide-ranging. But at this early stage, researchers cannot say with certainty what specific benefits will arise. They are essentially flying blind because they know very little about the human microbiome and its role in healthy and unhealthy individuals.
What does the future hold for the HMP?
The HMP is only the tip of the iceberg for understanding the diversity of the human microbiome and its role in healthy and unhealthy individuals. The next step is to broaden the human subject sampling. The 300 normal, healthy individuals who contributed to the reference microbiome had a mean age of 26, were all Americans, and were mostly white. Such a small and biased sample disregards the effects of age and cultural practices on the diversity of the microbiome. For example, it is known that diet influences the diversity of the gut microbiome and that humans are born sterile with no associated microbiome and begin accumulating a microbiome following birth. It then takes about two years for an infant’s microbiome to stabilize. It is unknown how (or if) the microbiome diversity changes as individuals age. These are all questions that still need to be addressed.
For more information
- http://www.sciencedaily.com/releases/2010/05/100520141214.htm
- http://commonfund.nih.gov/hmp/
- http://www.nih.gov/news/pr/dec2007/od-19.htm
- http://www.nih.gov/news/health/oct2008/nhgri-07.htm
- http://www.ncbi.nlm.nih.gov/bioproject/43021
- PLOS Series: http://www.ploscollections.org/article/browseIssue.action?issue=info:doi/10.1371/issue.pcol.v01.i13
The NIH Microbiome Cloud Project
A New Resource to Help Researchers Explore Microbiome Data

COURTESY: NIAID
The NIH Microbiome Cloud Project (MCP), led by the National Institute of Allergy and Infectious Diseases (NIAID) and the National Human Genome Research Institute (NHGRI), addresses one of the greatest challenges facing microbiome scientists: large-scale data analyses. A team of scientists from NIH, academia, and industry is developing a cloud, or Internet-based, platform that brings together Human Microbiome Project (HMP) data and analysis tools. The HMP produced 14 terabytes of genetic information about the microbes that naturally colonize our bodies. That’s enough data to fill more than 3,000 standard DVDs.
“The amount of data that can now be generated is orders of magnitude higher than what could be done just a few years ago,” said John Tsang, chief of the Systems Genomics and Bioinformatics Unit in NIAID’s Laboratory of Systems Biology. Tsang’s research focuses on understanding biological interactions such as those between the microbiota and the human body. He plans to use the cloud platform to analyze his laboratory’s experimental microbiome sequencing data and compare them with HMP data.
Mining microbiome datasets promises to help scientists better understand the role of the microbiota in health and disease and identify new targets for drugs and vaccines, but scientists need proper tools to make sense of these complex data. Many researchers do not have access to the high-performance computing resources, data-analysis tools, or the technical expertise required to assemble and study large-scale microbiome datasets. By bringing together the data and tools in the cloud, the MCP will give researchers access to vast amounts of data with high-performance computing power.
“A cloud platform that provides access to centralized data-analysis resources is a great step forward,” Tsang said. “The availability of an established set of tools used by multiple investigators will facilitate our ability to explore large datasets.”
In September 2013, NIAID and NHGRI launched the first phase of the MCP, which makes a five-terabyte portion of HMP sequencing data publicly available on the Amazon Web Services cloud. Cloud storage facilitates analysis by reducing the need for time-consuming data downloads. The MCP team is developing the next phase of the project, which will add analysis tools, more data, and supporting documentation such as online tutorials. Before the platform is released to the public, a team of NIH scientists and extramural researchers will evaluate it and provide feedback on its functionality and usability.
The MCP’s cloud environment promises to encourage greater collaboration and data sharing among NIH intramural and extramural researchers and their colleagues. Scientists’ experiences with the MCP also will help inform NIH best practices for using cloud technologies for biomedical research.
This page was last updated on Thursday, April 28, 2022
