New Software Program Evaluates Potential for Metastasis
NIEHS Scientist Leping Li Develops New Approach for Classifying Cancer Cells
Pathologists have traditionally used the physical characteristics of melanoma cancer cells to classify them as primary or metastatic. Recently, however, computational biologist Leping Li at the National Institute of Environmental Health Sciences (NIEHS) developed another approach to classifying the cancer cells.
Melanoma is the most serious type of skin cancer and develops in the pigment-producing melanocytes in the basal layer of the epidermis. Primary melanoma represents the original site of the tumor; the metastatic form means the tumor has spread and has become life-threatening.
Li has developed an algorithm, based on gene-expression data, that evaluates each tumor’s resemblance to metastatic tumors. He presented his work as part of the NIH Director’s Seminar Series, April 4, on NIH’s Bethesda campus.
LEPING LEE, NIEHS
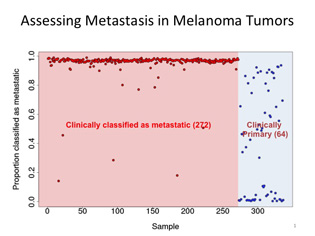
Leping Li (NIEHS) analyzed gene-expression data from 336 TCGA melanoma samples using the new GA/KNN software program. Nearly all of the 272 clinically classified metastatic tumors were scored as likely to be metastatic, while the 64 primary tumors separated into three categories: one-third had characteristics like metastatic tumors, one-third correlated with primary tumors, and one-third fell somewhere in-between.
It all started in 2000, when Li was a member of the NIEHS Laboratory of Structural Biology. He and several senior researchers wrote a software program called Genetic Algorithm/K-Nearest Neighbor (GA/KNN), a classification tool for categorizing genes found by microarray analysis (Bioinformatics 17:1131–1142, 2001).
Li offered the program on his Web site as freeware (http://1.usa.gov/1olDU75) and soon after joined the NIEHS Biostatistics Branch, receiving tenure in 2012.
In the fall of 2013, Li was examining The Cancer Genome Atlas (TCGA), a cancer database funded and managed by the National Cancer Institute and the National Human Genome Research Institute. As he looked at the gene-expression and mutation data for a variety of tumor types, he realized the tools he developed back in 2000 could be applied to the new, more accurate NextGen Sequencing data.
“What if the clinical classification says one thing,” Li began, “but the gene-expression data [say] something else?”
Li made several modifications to the GA/KNN software and decided to test it on melanomas because they are highly metastatic. He analyzed gene-expression data from 336 TCGA melanoma samples: 272 were clinically classified as metastatic and 64 as primary.
Ninety-eight percent of the 272 metastatic tumors displayed gene-expression patterns that were similar to one another. But to his surprise, nearly two-thirds of the nonmetastatic primary melanomas exhibited expression patterns that were typical of metastasis.
In addition, the updated GA/KNN revealed 39 genes that seemed to be taking part in metastasis. During the tumors’ switch from primary to metastatic, the genes’ expression levels either increased or decreased, suggesting that these genes take part in metastasis. Most of them are known to be involved in ectoderm and epidermis development. Several others haven’t been reported in the literature and form the basis of Li’s research.
“Our analysis may provide useful information for treatment and disease management for melanomas in the future,” Li said. “It may also offer insight into the molecular mechanisms that underlie metastasis.”
Although the newer version of GA/KNN isn’t publically available yet, Li’s software tweaks will give researchers another tool in the fight against cancer, said NIEHS Biostatistics Branch Chief Clarice Weinberg, who helped create the original computer program.
“Leping’s creative development of algorithms may provide important clues based on mining gene-expression data,” she said. They “could ultimately give clinicians a way to focus on the most dangerous cancers.”
To see a videocast of Li’s presentation, “Gene Selection and Sample Classification with Applications to TCGA Data,” at the April 4, 2014, NIH Director’s Seminar Series, go to http://videocast.nih.gov/launch.asp?18378. For more on Li’s work, visit https://irp.nih.gov/pi/leping-li.
This page was last updated on Wednesday, April 27, 2022
