Research Briefs
Read about scientific advances and discoveries by NIH intramural scientists: CAR T cells target solid tumors; CRISPR deletion scanning reveals protein function; stress influences social behavior; high-resolution thymic map added to Human Cell Atlas; and how living neurons and artificial intelligence systems filter input in similar ways.
NCI: CAR-T-CELL THERAPY SHOWS GREAT PROMISE IN GASTRIC AND COLORECTAL CANCERS
Advanced gastric and colorectal cancers are difficult to treat. The primary reason is that the cancer has spread to other organs, rendering traditional cancer treatments such as surgery and chemotherapy nearly futile. Thus, there is an urgent need to develop more effective therapies for these patients. Chimeric antigen receptor (CAR) T-cell therapy is a relatively new weapon in the anticancer arsenal, but it has mostly been successful against blood cancers. However, investigators at NCI led by Raffit Hassan and postdoctoral fellow Sameer Mir in the Thoracic and GI Malignancies Branch have taken a novel approach to extend CAR-T-cell therapy to the realm of solid tumors.
As detailed in a November 2024 Clinical and Translational Medicine article, Hassan’s team effectively targeted human gastric and colorectal cancers grafted into the bodies of specially-bred mice. Building on research by other groups, the team used a CAR T cell called hYP218, which is designed to recognize mesothelin, a cell-surface protein that is overexpressed in these types of cancers. T cells are a natural part of the immune system, and CAR T cells are essentially trained to kill cancer cells rather than viruses, bacteria, or other harmful agents in the body. The idea behind using hYP218 CAR T cells is that, because mesothelin is far more abundant on the surface of cancer cells, targeting it with these bioengineered immunotherapeutic T cells will kill the cancer while minimizing damage to healthy cells that have little to no mesothelin protein. The research team noted that activated hYP218 CAR T cells persisted in the tumor microenvironment and retained their cytotoxic activity.
Hassan’s team additionally found that these CAR T cells had an enhanced efficiency when combined with the drug pembrolizumab, particularly in mice with larger, more established tumors. Pembrolizumab is a checkpoint inhibitor that blocks the PD-1 pathway, a naturally occurring mechanism that suppresses T-cell activity.
“Based on the promising preclinical activity against different solid tumors, a phase 1 clinical trial of hYP218 CAR T cells will open at the NIH Clinical Center in 2025 for the treatment of patients with advanced mesothelin-expressing solid tumors who have failed standard therapies,” said Hassan, who will serve as the principal investigator of this clinical trial. (NIH authors: S. Mir, A. Venugopalan, J. Zhang, N.U. Nair, M. Sengupta, M. Khanal, C. Stathopoulou, Q. Jiang, and R. Hassan, PMID: 39548594)
NHGRI: CRISPR SCANNING METHOD REVEALS PROTEIN FUNCTION
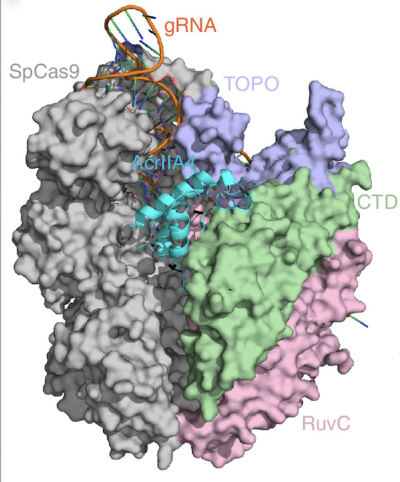
CREDIT: SYSTEMS BIOLOGY AND GENOME ENGINEERING SECTION, NHGRI
Shown in cyan is the structure of the AcrllA4 protein binding to the Cas9 enzyme (specific domains in green, lavender, and pink, with the remainder in gray). NHGRI scientists tested a new method that revealed just how AcrllA4 functions to foil bacteria’s CRISPR-Cas9 gene-editing machinery.
CRISPR technology, often associated with gene therapy, could be deployed to accurately and efficiently understand protein function, according to a new study by NHGRI scientists.
The researchers tested an approach known as deletion scanning on AcrllA4, a protein that bacteriophages (phages) encode to foil bacteria’s CRISPR-Cas9 immune defense. Phages and bacteria have been battling it out for eons, and many phages have evolved the ability to resist CRISPR-Cas9, a mechanism whereby bacteria incorporate a sequence of viral DNA into their own genome.
To uncover the workings of phage’s anti-CRISPR adaptation, the investigators, somewhat ironically, used CRISPR to comprehensively generate thousands of genetic deletions affecting specific regions of AcrllA4. They were then able to test the effect of each modification on blocking the activity of Cas9, an enzyme that cuts DNA. Their high-throughput data showed that AcrllA4 acts upon Cas9’s ability to bind to a short sequence of DNA used by the enzyme to recognize target sites just before cutting the DNA strand.
Beyond working out the function of AcrllA4, the authors note that such comprehensive deletion scanning could have many applications. Those include engineering new or improved proteins, revealing evolutionary adaptations, and understanding how deletion variants contribute to genetic diseases. (NIH authors: A.B. Iturralde [now at University of Virginia, Charlottesville], C.A. Weller, S.M. Giovanetti, and M.J. Sadhu, PMID: 39570312)
NIEHS: BRAIN RESPONSE TO SOCIAL STRESS INFLUENCES LATER BEHAVIOR
Stressful experiences can alter how an individual responds to future social interactions. A study led by NIEHS researchers suggests that the effects of stress on social behaviors depends on how resilient the brain is during those experiences. By exposing mice to a stressful social experience—being socially defeated by a more aggressive, dominant mouse—the investigators examined the neurobiological processes that differ between those who are more and less susceptible to stress-driven behavioral changes.
Avoidant behaviors and reduced social exploration were evident in mice that experienced social defeat and persisted for weeks after the event. Moreover, mice that exhibited defensive behavior during the defeat were more resilient and were more likely to socialize later than the mice who fled the stressful encounter. This effect was found to be driven by activity in the cornu ammonis 2 (CA2), a subregion of the hippocampus that is critical for social learning and aggression in mice.
The scientists used viral targeting of molecules to selectively silence CA2 neurons during acute social stress and to trace the downstream projections of these neurons. Experiments revealed that decreasing CA2 activity enhanced avoidant behaviors after social stress, suggesting that activation of the CA2, and its downstream targets in the hippocampus and cortex, promotes resilience and reduces behavioral sensitivity to acute social stress. (NIH authors: D. Radzicki, K.E. McCann, G.M. Alexander, and S.M. Dudek, PMID: 39548322)
[BY ASHLEY PRATT, NICHD]
NIAID: HIGH-RESOLUTION MAPPING OF HUMAN THYMUS ADDED TO THE HUMAN CELL ATLAS
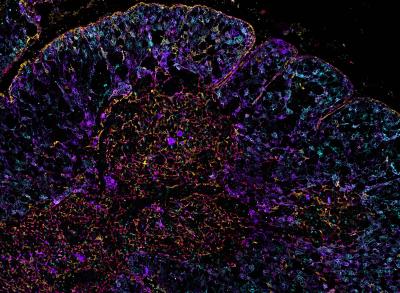
CREDIT: ANDREA RADTKE, RONALD GERMAIN, NIAID LABORATORY OF IMMUNE SYSTEM BIOLOGY
Iterative bleaching extends multiplexity (IBEX) image of the human thymus. Thymic epithelial cells are labeled with DEC205 (cyan), pan-cytokeratin (purple), keratin 5 (red), and keratin 14 (yellow).
Researchers at NIAID and their international colleagues developed a novel mathematical model that combines highly multiplex, high-resolution optical imaging, single-cell RNA sequencing, and spatial transcriptomic data. Termed OrganAxis, the model can be applied to any tissue, and when applied to the human thymus, the scientists produced the most detailed description of the organ to date. The thymus is a vital immunological organ that supports the maturation of T cells. Disorders affecting the thymus can result in devastating autoimmune diseases, immunodeficiencies, and cancers.
By combining technologies, including NIAID-developed iterative bleaching extends multiplexity, known as IBEX (PMID: 33376221), the researchers established a framework called the corticomedullary axis. They applied that framework to map the cellular landscape of the human thymus in exquisite detail, including the trajectories of immune cells (thymocytes) as they pass through the organ during maturation and selection together with the signaling gradients that guide them. “The common coordinate framework established here empowers integration of diverse datasets, providing a strategy for charting thymic changes using new single-cell and spatial technologies,” said Andrea Radtke, a senior author on the study.
According to Ronald Germain, fellow author and NIH distinguished investigator, combining both high- and low-resolution technologies revealed how valuable such imaging can be in creating very precise maps of a tissue. “Our ongoing methods are now extending such imaging to 3D, which will produce true organ-level maps in the near future,” he said.
The thymic atlas adds to a global collaborative attempt to map every cell in the human body, collectively termed the “Human Cell Atlas.” (NIH authors: B.T. Wachter, R.T. Beuschel, M. Bosticardo, F. Pala, L. Notarangelo, R. Germain, and A.J. Radtke, PMID: 39567784)
[BY TAYLOR FARLEY, NIAID]
NIMH: OPTOGENETICS REVEALS HOW NEURONS FILTER INPUT IN VIVO
How information flows in living neurons may share similarities with how information is processed by artificial intelligence (AI) systems, according to a study by NIMH scientists. Using new techniques, they measured activation functions, that is, the firing rate produced for a given input level, of the neurons of active brains in awake mice for the first time.
The researchers used an advanced method called two-photon holographic stimulation, which uses two high-powered lasers to image and stimulate cells at the same time. The technique relies on optogenetics technology, which modified the brain cells to activate in response to light. Neurons were stimulated in combination with visual input, allowing the team to collect data from hundreds of neurons in various states of activity.
They found nearly linear responses when neurons’ activity level was elevated, and in contrast, significantly attenuated responses when neurons’ activity was suppressed. This result shows that living neurons can, by changing activity level, filter out certain inputs while prioritizing others.
According to the authors, the findings suggest that both brains and AI systems face similar selection pressures—perhaps as the result of the need of both to learn—and this activation function shape is a common and shared solution. (NIH authors: P.K. LaFosse, Z. Zhou, J.F. O’Rawe, N.G. Friedman, V.M. Scott, Y. Deng, and M.H. Histed, PMID: 39485801)
This page was last updated on Friday, January 3, 2025
