Unfolding the Mystery of Transformer-like Proteins
Lauren Porter’s Research on Fold-switching Proteins
For decades it was thought that when proteins fold, they assume only one stable structure that performs a specific function. For example, the unique structure of the protein hemoglobin allows it to transport oxygen from the lungs to the tissues. This one-sequence-one-fold paradigm is attributed to NIH’s Christian Anfinsen, who won the 1972 Nobel Prize in Chemistry. His work established a connection between the amino acid sequence that makes up a protein and its three-dimensional shape, or conformation, that dictates the protein’s biological function.
CREDIT: CHIA-CHI “CHARLIE” CHANG, OD
Lauren Porter’s research focuses on fold-switching proteins, many of which are associated with diseases such as cancer, autoimmune disorders, and bacterial and viral infections.
Stadtman Tenure-Track Investigator Lauren Porter’s research on fold-switching proteins challenges the ubiquity of this one-sequence-one-structure paradigm. “Fold-switching proteins are like Transformers, like Optimus Prime,” said Porter, referring to the hero of the science fiction franchise of shapeshifting humanlike robots. “Sometimes he’s a robot, and sometimes he turns into a car. He uses both of his structures and both of his functions to fight crime.” Similarly, the proteins she studies have multiple stable structures and functions.
During her college days at the University of Maryland Baltimore County (Baltimore), when Porter majored in physics and math, her dad was diagnosed with stage IV lymphoma. Seeing him go through rounds of chemotherapy influenced Porter to pursue her Ph.D. in biophysics at Johns Hopkins University (Baltimore), where she hoped she could use her knowledge to contribute to better medical treatments.
During her graduate training, she became interested in fold-switching proteins and their association with cancer and other diseases. She pursued that interest through her postdoctoral training and as a research scientist at the Howard Hughes Medical Institute’s Janelia Research Campus (Ashburn, Virginia) before joining NIH in 2019 as a tenure-track Stadtman Investigator with a primary appointment at the National Library of Medicine (NLM) and a secondary one at the National Heart, Lung, and Blood Institute. She is developing methods that predict fold switching from genomic sequences. Her team validates those predictions using circular dichroism (a form of spectroscopy that uses polarized light to study the structure of molecules), and characterizes the properties of fold-switching proteins.
Brian Volkman, researcher and biochemistry professor at the Medical College of Wisconsin (Milwaukee, Wisconsin), reported one of the first examples of a fold-switching protein in 2008, with the case of human chemokine lymphotactin. This immune protein can exist in two different conformations under normal physiologic conditions. Since then, about 100 fold-switching proteins have been identified, but it is difficult to know just how common they are. There are about 220 million unique protein sequences and scientists have only identified the protein structure for about 58,000 of them. Current methods, such as the algorithm AlphaFold2, predict protein structure based on correlations with protein sequence but are unable to predict fold switching.
“Clearly there’s some fundamental biophysics we don’t understand,” said Porter. “Our research is really trying to understand the physical and chemical underpinnings of protein folding and fold switching.”
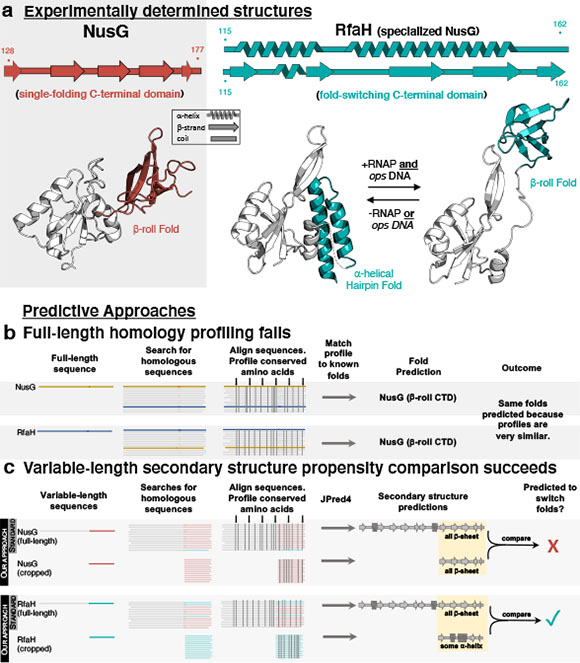
CREDIT: bioRxiv
Figure 1. (a) An example of fold-switching: RfaH’s C-terminal domain (teal) completely switches between alpha-helical and beta-sheet folds, while the C-terminal domain of its homolog, NusG (red) maintains a b-sheet fold. An overview of the methods used to predict protein structure (b) and fold-switching (c) are shown below.
Porter approaches her work by looking at these proteins from as many perspectives as possible, such as using high-throughput analysis to evaluate how good their predictions are and when they fail. Her group will soon begin to study protein dynamics by analyzing the intermediate steps of fold switching to get a clearer picture of how these proteins are able to change their three-dimensional structures. Another method to better understand fold switching is mutational analysis, in which multiple sites on the protein are mutated and researchers observe which mutations hinder the protein’s ability to switch conformations.
Fold switching seems to be a specific mechanism of regulation that enables cells to respond to their environment very quickly. Having a protein that already exists switch to a different form is a faster and more efficient form of regulation than transcribing and translating a whole new protein. Some of these proteins will exist in both forms at equilibrium, whereas others switch forms in response to specific triggers such as changes in pH or temperature.
A number of fold-switching proteins are associated with diseases such as cancer, autoimmune disorders, and bacterial and viral infections. However, without fully understanding how these mercurial proteins work, it is difficult to make targeted treatments.
“My hope is down the road, therapeutics could be developed that target fold-switching proteins and force them to stay in one conformation or favor one conformation,” said Porter. “If by the end of my lifetime there was even one therapeutic based on this, that would be amazing.”
When Porter began her work on fold-switching proteins, not everyone was convinced they were widespread or important. Porter’s advice for trainees is to follow the research that interests them the most, even if others are unsure of where it may lead.
To learn more about Porter’s research, you can watch a videocast of a presentation she gave at the National Library of Medicine Virtual Town Hall on January 19, 2022, at https://videocast.nih.gov/watch=44410 (NIH only). Her 15-minute talk, titled “What Proteins and Optimus Prime Have in Common,” begins about 23 minutes in.

Natalie Hagen is a postbaccalaureate research fellow in the National Center for Advancing Translational Sciences, where she is performing pharmacokinetics studies of novel drug candidates. Upon completion of her fellowship in 2022, she will be pursuing a Ph.D. in molecular biology at the Perelman School of Medicine at the University of Pennsylvania (Philadelphia). In her spare time, she likes to run, read, and go hiking with friends.
This page was last updated on Tuesday, May 17, 2022
