Today’s Discoveries, Tomorrow’s Cures
Report From the 2012 Research Festival
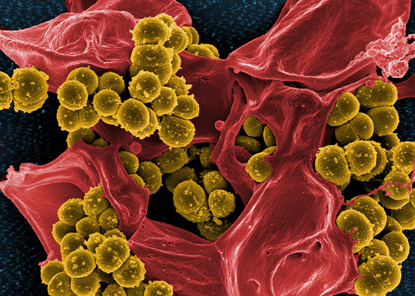
FRANK DELEO, NIAID
Methicillin-resistant Staphylococcus aureus (MRSA) bacteria (yellow ball-like structures) are bursting out of a dead neutrophil. This scary image serves as a reminder of our past battles with epidemic diseases such as cholera, plague, and smallpox, as well as our current challenges with emerging threats such as MRSA.
Only one event brings together the intramural community year after year, and that’s the NIH Research Festival. What began as “NIH Research Day” on September 25, 1986, has evolved into a four-day festival featuring scientific talks, posters, exhibits, and more.
“It’s gotten bigger and better every year,” said NIH Director Francis Collins. “This year is the biggest ever.”
This year’s festival featured a unique plenary program, concurrent symposia with 26 topics and 148 talks, more than 500 posters, special exhibits, and a scientific equipment tent show. There was also a special session with U.S. Congressman Andy Harris (Maryland), a trained physician and recipient of NIH grants, who talked about the importance of NIH’s biomedical research.
The festival took place October 9 through 12 and merged with the NIH National Graduate Student Research Conference, giving 120 graduate students studying at U.S. universities the chance to present their work to intramural scientists and learn about research going on at the NIH.
The 2012 Research Festival was particularly momentous because it marked NIH’s quasquicentennial, or 125th anniversary. Four institutes also achieved milestones this year: It is the 10th anniversary of the National Institute of Biomedical Imaging and Engineering (NIBIB), the 50th for the National Institute of Child Health and Human Development (NICHD) and the National Institute of General Medical Sciences (NIGMS), and the 75th for the National Cancer Institute (NCI).
The theme of the 2012 festival, “The NIH at 125: Today’s Discoveries, Tomorrow’s Cures,” was reflected in the festival artwork used on posters, program booklets, and the Web site. Created by Frank DeLeo’s group at the National Institute of Allergy and Infectious Diseases (NIAID), it depicts methicillin-resistant Staphylococcus aureus (MRSA) bacteria bursting out of a neutrophil. This scary image serves as a reminder of our past battles with epidemic diseases such as cholera, plague, smallpox, and yellow fever, as well as our current challenges with emerging threats such as MRSA.
The plenary session included three dynamic big-vision talks that featured eminent cutting-edge research within the intramural program and proposed future applications for research at the NIH. “The plenary session was fantastic because it captured the very essence of NIH intramural programs,” said Antonello Bonci (scientific director in the National Institute on Drug Abuse), who co-chaired the event with Constantine Stratakis (scientific director at NICHD). “It was diverse in scientific interests and disciplines,” Bonci continued. “Cutting-edge and highly innovative yet very much in synergy with each other.”During his opening remarks at the plenary session, Collins encouraged intramural scientists to use the Research Festival as an opportunity to interact and trigger collaborations with researchers at other institutes. “We all have our own circles that we mix and mingle with in terms of our research interests,” said Collins, “but here’s a chance to enlarge your circle in a much bigger and broader way…to really expand your horizons.”
“It was our very own voyage of the Intrepid, led by four fearless NIH explorers, past and present,” said Stratakis.
Gary Gibbons described his research on the etiologies of racial health disparities in cardiovascular disease (CVD) using a multilevel biosocial approach. His group conducted a study that found that obesity prevalence was a key driver in CVD. Certain lifestyle determinants, including lower physical activity and lower fruit and vegetable intake, serve as drivers for obesity. “We recognized that where you live, work, and play [has] an impact on your health,” said Gibbons, who’s the director of NHLBI. While studying health maladies from a social context is provocative, it is now supported by research that demonstrates how behavior permeates through a social network. Ultimately, biosocial systems may predict who is at risk for a particular disease and to design preemptive treatments long before clinical symptoms develop.
Jennifer Lippincott-Schwartz (NICHD) studies cellular organization and function using photoactivatable fluorescent proteins and a technique she helped develop at NIH, known as photo-activated localization microscopy (PALM). Unlike conventional microscopes that have a best resolution of 250 nanometers, a PALM setup has an X-Y resolution of 20 nanometers, which creates sharp super-high-resolution images. Lippincott-Schwartz pointed out “how important technology is in order for us to continually move forward in our discoveries about basic scientific processes as well as diseases.” She explained how navigating the cellular landscape with new optical probes and imaging strategies, allows her group to quantify and monitor protein and organelle turnover in cells and determine how molecules are organized and clustered for signaling within the plasma membrane.
Ron Germain (NIAID) uses multiphoton intravital imaging to study the role of anatomy in the operation of the immune system. When tissue is damaged, neutrophils rapidly congregate in the region of damage. “But how do these cells know that there is damage there and to [go] into this region?” Germain the audience. He explained how his research shows that integrin-mediated adhesion is essential for neutrophil migration to the central lesion.
Germain is also interested in what keeps pathogens from spreading in the lymph before the adaptive immune system kicks in. His group found that macrophages in lymph nodes are critical in preventing bacterial movement into the lymphatic system. His work also indicates that the lymph node is a site of adaptive immunity, where lymphocytes are localized to the periphery near pathogen entry portals. These fantastic voyages into the heart of the immune system will drive future multiscale approaches in predictive immunology.

ERNIE BRANSON
Joseph James Kinyoun, the founder of the precursor to NIH in 1887, made an appearance at the 2012 Research Festival to address the things that continue to haunt him 93 years after his death. Kinyoun was played by intramural scientist David Robinson (NIAID) who enjoys being an actor in his spare time.
To celebrate the history of NIH, a very special guest addressed the audience next. Joseph James Kinyoun was the founder and first director of the Marine Hospital Service Laboratory of Hygiene in 1887. Although Kinyoun died in 1919, he made an appearance at the 2012 Research Festival to address three things that continue to haunt him 93 years after his death. “I have been resting uneasy with unfinished business,” announced Kinyoun (played by David Robinson, assistant scientific director at NIDCD), who referenced several diseases that were rampant during his lifetime, the need for the transfer of organs to extend human life, and the need for federal investment in a national research enterprise. Deputy Director of Intramural Research Michael Gottesman assured Kinyoun that he could rest easy knowing that many of those diseases have since been eradicated, that the technique of organ transplantation is in use today, and that in 1930 Congress renamed Kinyoun’s Laboratory of Hygiene the National Institute of Health. A 64-page booklet of Kinyoun’s legacy, The Indispensable Forgotten Man, written by David Morens and Anthony Fauci, can be downloaded from the festival Web site (https://researchfestival.nih.gov/2012/kinyoun.pdf). An article about Kinyoun also appears in this issue of the Catalyst.
The plenary session ended with a panel of three distinguished intramural investigators—William Paul, Judy Rapoport, and Stephen Katz—who discussed their experiences at NIH and why they choose to do their research here.
William Paul (NIAID), who first came to NIH as a medical student in 1959, pointed out that there is a sense of noncompetitiveness in the intramural community that doesn’t exist in other research institutions. “I can count on my colleagues to want my work to succeed and to do everything they can do to help it,” said Paul. “And I feel the same way [about their work].”
Judy Rapoport (NIMH) was one of the first investigators to start child psychiatry research at NIH. One of the strengths of working at NIH, according to Rapoport, is the ability to study rare disorders such as childhood schizophrenia. “It’s been the sense of an imaginative, flexible, and very empathic administration, together with other things easing the road for clinical research, that has made it very hard to consider moving anywhere else.”
Stephen Katz (NIAMS), who came to NIH in 1974 as a principal investigator in dermatology, said that part of what makes NIH such a great place to work is the ability to join clinical practice with basic science. “We have time here,” said Katz. “We have time for patients, we have time for research, and we have time for collaborations.”
What follows is a snapshot of Festival sympsia.
SELECTED SYMPOSIA
Using Chemistry to Unravel Biology
Chemists make the world go round … in the way that their work helps scientists understand biology in action.

MICHAEL BEWLEY
Carole Bewley (NIDDK) shown collecting rare Chrysophaeum taylori algae when she was scuba diving in the Virgin Islands. In the lab, she extracted chrysophaentin A, a natural compound that killed every gram-positive drug-resistant organism it came into contact with.
To understand the biology of addiction, Amy Newman (NIDA) uses chemistry to develop fluorescent tropane-based tools to visualize monoamine transporters in action in the central nervous system. Cocaine, a highly addictive drug that binds to all three monoamine transporters, is a tropane-based molecule and provides the template for these tools. Newman confessed that 20 years ago, when she was doing her postdoctoral training in NIDDK, she published research on another fluorescent compound she had developed, but her paper was never cited.
“I’d like to think we were ahead of our time,” she told the chemistry fans who had gathered to hear her talk. But today, her papers are cited aplenty. Her work demonstrates that the combination of state-of-the-art synthetic organic chemistry techniques with molecular modeling and interpretation of pharmacological data has resulted in the discovery of important molecular probes for studying neurochemical targets. She hopes this multidisciplinary approach will provide new leads toward the development of pharmacotherapeutic agents that can be used to treat addiction.
To understand the biology of antibiotics, Carole Bewley (NIDDK) uses chemistry to trap anti-infective targets of marine natural products. She told a story of how in 2007, she was snorkeling with her family in the Virgin Islands when she found a fluffy yellow organism. She gathered some samples, took them back to her NIH lab, and extracted what she thought was a cyanobacterium. But the isolate turned out to be a rare alga that produced chrysophaentin A, a natural compound that killed every gram-positive drug-resistant organism—such as methicillin-resistant Staphylococcus aureus—it came into contact with. With the help of the postdocs and a graduate student in her lab, Bewley determined that the compound inhibits the bacterial cytoskeletal protein FtsZ, an enzyme necessary for the replication of bacteria. Bewley’s discovery has been patented by NIH; NIDDK is looking for collaborators to further develop, evaluate, or commercialize the chrysophaentin antibiotics.
Many other NIH chemists are developing and using molecular tools to help their colleagues—chemists and nonchemists alike—gain a better understanding of biology.
From the 2012 Research Festival’s session “Molecular Tools; Using Chemistry To See, Wrestle, Unravel, and Trap Biology,” chaired by Amy Newman (NIDA) and Dan Appella (NIDDK); held on October 10, 2012.
Quantitative Biology at the Single-Cell Level
Whether individual cells make up the developing brain, the salivary gland, or a cancer mass, each has a unique story of growth, migration, and signaling. NIH researchers use a variety of live-cell imaging techniques to study real-time changes in individual cells.
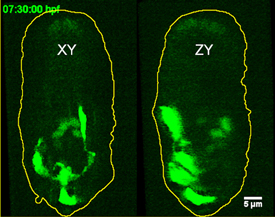
YICONG WU, NIBIB
Hari Shroff used iSPIM, an advanced imaging technique, to view neurons migrating in the developing roundworm embryo. Lateral (left) and axial (right) fluorescent images are depicted seven and a half hours into embryogenesis.
What’s important, said Hari Shroff (NIBIB), is figuring out how to capture images of cells without killing them. “Usually, if you try to increase the resolution or try and go faster, you end up destroying the cell, also faster.” His solution is inverted microscope selective-plane-illumination spectroscopy (iSPIM), a technique in which a thin sheet of light illuminates a single plane within a three-dimensional specimen. Using iSPIM, Shroff captures images of neurons as they grow and migrate within live, developing Caenorhabditis elegans (roundworm) embryos.
Myong-Hee Sung (NCI) takes a different approach. She uses mathematical modeling and an optical imaging technique called fluorescence recovery after photobleaching (FRAP) to study the dynamics of the transcription factor nuclear factor–kappa light-chain-enhancer of activated B cells (NF-kappaB) as its activity oscillates on and off. By using single-cell imaging, Sung reveals continuing asynchronous oscillations of NF-kappaB activity in individual cells, oscillations that dictate unique transcription events. And with new technologies, such as temporal image-correlation spectroscopy, which improves upon conventional FRAP, Sung is beginning to study the spatial heterogeneity of NF-kappaB within the nucleus itself.
From the 2012 Research Festival’s session “Quantitative Biology at the Single Cell Level,” co-chaired by Eric Batchelor (NCI) and Myong-Hee Sung (NCI) and held on October 9, 2012.
Beyond the Fig Leaf
Biomedical scientists are beginning to realize that they must take sex and gender into consideration when studying diseases or developing treatments and therapeutic devices. Sex (male or female) and gender (behavioral, cultural, or psychological traits associated with one sex) are terms used to categorize similarities and differences in perception, experience, treatment, and outcome of disease.
Cardiologist Nakela Cook (NHLBI)presented staggering data on the prevalence of heart disease in women. Cardiovascular disease (CVD) kills about 400,000 women each year—more than cancer. Each year, about one in 31 female deaths is from breast cancer, but one in four is from heart disease. Yet there is an underperception of risk and an under-recognition of symptoms in women. Smoking, obesity, and high blood pressure put both sexes at risk for heart disease, but in women other risk factors include pre-eclampsia (hypertension in pregnancy), gestational diabetes, and a total cholesterol concentration of over 200 milligrams per deciliter of blood.
Men and women have different symptoms, too. Both may experience chest pain during a heart attack, but a man may feel a vise-like pressure whereas a woman may feel a stabbing, pulsating, or sharp pain. Women are also more likely to report nausea, shortness of breath, or abdominal pain—unexpected symptoms that can lead to misdiagnosis and delays in treatment.
Compounding the problem is that clinical trials for CVD therapies and devices have tended to enroll fewer women than men and have failed to report sex-specific results. Cook emphasized the need for an increased enrollment of women in trials for heart disease.
The battle of the sexes is not over when it comes to pain, according to Women and Sex/Gender Differences Research Coordinator Cora Lee Wetherington (NIDA). Pain disorders are more prevalent in women than in men, including pain disorders of the head (migraines are more common in women and cluster headaches in men), neck, shoulder, hips, mid-abdominal, finger joint, and mandibular areas. In addition, more women than men suffer from fibromyalgia, rheumatoid arthritis, osteoarthritis, and multiple sclerosis.
Women’s increased sensitivity to pain may be attributable to differences in the nervous system’s processing of pain, sex hormones, genetics, gender roles, mood, emotions, and stress and coping mechanisms. Women even respond differently to treatments for pain. Research on sex-differences data analysis in animal-models and better studies that include women could provide clues to effectively managing chronic pain in both men and women, said Wetherington. And that’s good news for everyone.
From “Beyond the Fig Leaf: The Science of Sex and Gender Differences,” chaired by Janine Clayton (Director, ORWH) and held on October 9, 2012. To see a videocast, go to https://videocast.nih.gov/launch.asp?17607
One Gene, One Disease, Two Races: A Transatlantic Journey
Given the way humans migrate throughout the world, few ethnic groups are completely free of genetic mixing. NIH researchers are trying to understand how ethnic differences—and the underlying genetics—may play a role in noncommunicable diseases.
Cheryl Winkler (NCI’s Frederick National Laboratory for Cancer Research) and Jeffrey Kopp (NIDDK) are using admixture mapping to trace genetic traits in people of mixed ancestry such as African Americans. Compared with European Americans, African Americans have a higher risk of developing chronic kidney disease (CKD).
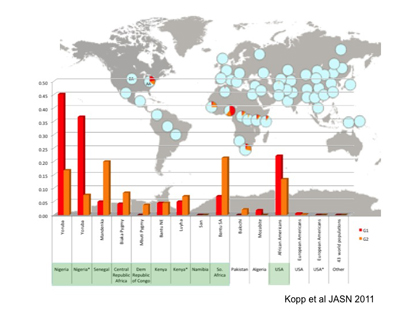
Worldwide frequency distribution of APOL1 G1 (red) and G2 (orange) alleles. Genotypes of G1 and G2 are determined for 51 populations in the Human Genome Diversity Panel (HGDP), for the HapMap Luhya population from Kenya (International HapMap Project [Phase II]), and for African Americans (AA) and European American (EA) controls in the NIH FSGS (Focal and Segmental Glomerulosclerosis) study cohort. The allele frequencies of G1, G2, and wild-type alleles (light blue) in each population are depicted in pie charts overlaid upon a world map. Allele frequencies for G1 and G2 for each population are shown in the chart at the bottom.
Winkler and Kopp and collaborators at Harvard Medical School (Boston) found that two sequence variants (G1 and G2) of the apolipoprotein-1 gene (APOL1), on chromosome 22, increased the risk of the kidney diseases focal segmental glomerulosclerosis, human-immunodeficiency-virus-associated nephropathy, and hypertension-attributed kidney disease. Recently Winkler reported on a strong association between CKD and APOL1 variants in African Americans with hypertension: Those with two risky variants of the gene are much more likely to develop progressive kidney disease (Kidney Int DOI:10.1038/ki.2012.263).
The APOL1 variants are not all bad, however. They are also key to the host defense against Trypanosoma brucei, the parasite responsible for African sleeping sickness. A single copy of the renal risk allele protects against sleeping sickness whereas two copies increase the risk for kidney disease. In the United States, 46 percent of African Americans carry one of the two APOL1 risk variants and 11 to 15 percent carry both.
On the other side of the Atlantic, these variants—especially G1—occur more often in West Africans. APOL1 risk variants also occur frequently in populations with recent African ancestry and are absent in other populations. Winkler and colleagues hypothesize that the risk variants, while critical for survival in Africa, may contribute to the high rates of renal disease in African Americans.
Winkler will continue to investigate the relationship of APOL1 risk alleles as well as the molecular mechanisms of the ApoL1 protein in CKD. She hopes her work will lead to improved diagnostic and therapeutic options for people with kidney disease. She and her colleagues will explore whether there is a role for APOL1 genetic testing in personalized medicine to identify individuals at particular risk for kidney disease and if so, to initiate periodic health screening and early therapy.
From the 2012 Research Festival’s session “Health Disparities: Advances in Translational, Clinical, and Population Sciences,” co-chaired by Anil Wali (NCI) and Jeffrey Kopp (NIDDK); sponsored by the NIH Translational Research Interest Group and held on October 10, 2012.
Understanding Parkinson Disease
Parkinson disease (PD) is an insidious neurodegenerative disease that affects more than one million older adults in the United States and destroys dopaminergic neurons within a region of the midbrain known as the substantia nigra. PD causes tremors, slowed movement, rigid muscles, difficulty walking, and changes in speech. At the first noticeable sensation of motor impairment, up to 80 percent of these neurons have already succumbed to the disease. Researchers such as Honglei Chen (NIEHS) understand the importance of developing a means to detect PD early. He is using data from several large population studies to track patients with clusters of premotor symptoms such as loss of smell, constipation, and sleep disturbances. None of these symptoms guarantee that an individual will develop PD, but Chen hopes to identify and predict who is at risk for the disease and to understand the early disease etiology.
Postdoctoral fellow and Fellows Award for Research Excellence (FARE) winner Sarah Rothman (NIA) is also unraveling the mysteries of PD. She is focusing on a mutation in the gene that codes for the protein alpha-synuclein, which makes up Lewy bodies (abnormal aggregates of protein) that develop in dopaminergic neurons in PD patients. She noticed that mice with the mutation did not gain weight even when fed a high-calorie diet. Rothman then reviewed published clinical literature on human PD patients and learned that certain patients experienced weight loss and a lower rate of diabetes. Rothman went back to her mouse studies and confirmed that PD mice fed a high-calorie diet expended more energy, showed better insulin regulation, and had less body fat than normal mice fed the same high-calorie diet. She explained that a possible mediator of this effect might be decreased concentrations of leptin—a hormone responsible for suppressing appetite—in the mouse model and in people with PD.
From the 2012 Research Festival’s “Translational Research of Aging” session, co-chaired by Francesca Macchiarini (NIAID) and Ron Johnson (NCI); sponsored by the Geroscience Interest Group, and held on October 10, 2012.
The Perils of “NO” Dysregulation
The role of nitric oxide (NO) in malaria and sickle cell disease stole the show as one of the hot new research topics featured at the 2012 Research Festival. In healthy individuals, NO is a vasodilator that promotes vascular homeostasis and proper blood flow. But different conditions disrupt normal NO function, impeding its production or siphoning it away from its everyday targets.
The malaria and sickle cell research teams are working together to understand how an acute infection (malaria) and a chronic genetic disorder (sickle cell) cause disruption in blood flow to vital organs. They hope to test new therapeutics to restore red blood cell and vessel-wall function.
Matthew Alkaitis, a graduate student in Hans Ackerman’s lab (NIAID), does research on NO depletion during malaria. Malaria is caused by Plasmodium parasites, which are transmitted by certain mosquitoes. Symptoms of severe malaria are linked to vascular dysfunction—blood vessels become stressed and constricted, the parasites cling to vessel walls, and red blood cells have a harder time squeezing through small capillaries. These problems appear to have their roots in disrupted NO signaling.
Normally, NO formation depends on both the amino acid arginine and the cofactor tetrahydrobiopterin (BH4). While it is known that arginine concentrations drop in people with malaria, Alkaitis is examining an apparent reduction in BH4 as well. He is exploring whether BH4 could be a tool for restoring healthy NO signaling and stabilize the vascular system of infected patients while they undergo antiparasite treatments.
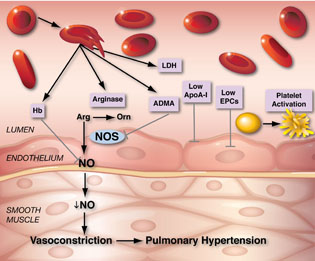
GREGORY KATO, NHLBI
In sickle cell disease, when red blood cells burst, one result is the release of free hemoglobin (Hb), that aggressively binds up nitric oxide (NO), stealing it away from its normal targets. Life-threatening problems such as pulmonary hypertension can result.
Gregory Kato (NHLBI) studies NO in the context of sickle cell disease, a genetic disorder causing crescent-shaped red blood cells that are rigid and fragile. When these cells rupture, free-floating hemoglobin spills into the bloodstream and binds up NO, stealing it away from its normal targets. The shortage of NO appears to contribute to the development of pulmonary hypertension and possibly leg ulcers. Kato has been pioneering ways to study and prevent these types of blood-system disruptions in sickle cell disease in order to shed light on associated life-threatening complications.
From the 2012 Research Festival’s session “Common Molecular Mechanisms Underlying Pathogenesis and Treatment of Human Diseases,” co-chaired by Minkyung (Min) Song (NCI) and Joel Moss (NHLBI); sponsored by the Translational Research Interest Group, and held on October 11, 2012.
Cancer Stem Cells (Almost) in a Dish
Cancer stem cells (CSCs) are enigmatic players in the cancer field, and some scientists even question their existence. But Tom Misteli’s lab (NCI) has created something that behaves a lot like a CSC, and this feat has many in the field excited.
Paola Scaffidi, a staff scientist in NCI’s Cell Biology of Genomes Group, which Misteli heads, has generated human cells with CSC properties in a culture dish by reprogramming somatic cells. Her work has demonstrated, for the first time, that somatic cells still possess enough plasticity to acquire CSC properties through oncogenic manipulation and may be the origin of some cancers.
This work also may provide a controlled system to study the biology of CSCs, which has been hampered by scientists’ inability to isolate pure CSC populations and manipulate them ex vivo.
According to the CSC hypothesis, cancers are hierarchically organized; a subset of cells, the CSCs, drives cancer growth. These CSCs are defined by their ability to initiate tumors and maintain their growth, to self-renew, and to differentiate into heterogeneous, nontumorigenic cancer cells.
Scaffidi and Misteli pulled off this trifecta with human skin fibroblasts. They built upon the work of the Bob Weinberg lab at the Massachusetts Institute of Technology (Cambridge, Mass.) by stably expressing human telomerase reverse transcriptase (hTERT), the oncogenic HRASV12 mutant protein, and the simian vacuolating virus 40 large T- and small T-antigens.
Their process transformed a subpopulation of fibroblasts into a more primitive, multipotent cell type that possessed all the hallmarks of CSCs and subsequently generated hierarchically organized tumors when injected into immunocompromised mice.
Much of this work was performed last year and described in a Nature Cell Biology paper (Nat Cell Biol 13:1051–1061, 2011). Scaffidi said she has since begun to characterize genetic and epigenetic events associated with CSCs’ tumorigenicity with the goal of uncovering new mechanisms of tumorigenesis and therapeutic targets.
These cells might be the first step toward understanding CSCs, along the lines of the aphorism scrawled on famed physicist Richard Feynman’s chalkboard, “What I cannot create, I do not understand.” Look for Scaffidi’s grand rounds talk on November 16, 2012, at which she will further explain her most recent work.
From the 2012 Research Festival’s session “Stem Cells in Development and Disease,” chaired by Steven Hou (NCI) and held on October 10, 2012.
Matrix Biology and Remodeling
Extracellular matrices (ECMs)—various interlocking meshes in which cells are embedded and that are composed of fibrous proteins, proteoglycans, and other molecules—make up the scaffolding that supports our cells, segregates tissues, and regulates intercellular communication. A well-functioning ECM is essential for development, morphogenesis, wound healing, and other physiological processes. But a misregulated matrix can cause damage ranging from macular degeneration, to arthritis, to tumor metastasis.
To fully understand how the ECM works, scientists need to study it in as natural an environment as possible. The trouble is that, until recently, most research was based on studies in conventional two-dimensional (2-D) cell culture. Kenneth Yamada (NIDCR), however, has been promoting the use of cell- and tissue-culture systems that mimic the natural ECM. His research team explored how matrix dimensionality (a fiber, a flat surface, or a gel, referred to as 1-D, 2-D, and 3-D, respectively) and matrix type (collagen, fibrin, or cell-derived matrices) regulate cell behavior.
Each different dimension and type of ECM tested had distinct effects on cell signaling and migration. For example, in 2-D environments, the signaling activity of Rac and other proteins at the front of the cell direct it to migrate forward. But Yamada’s team recently discovered that, in a 3-D cell-derived ECM, the same signaling is not required for efficient cell migration. Consequently, scientists need to rethink how cell migration is regulated in 3-D environments.
Yamada then expanded on the recent concept that ECM molecules can have major signaling roles beyond their structural functions, by describing two examples. The ECM proteins fibronectin and anosmin turn out to have distinct, dramatic effects on cell signaling, expression of specific genes, and even on the activities of individual growth factors during embryonic morphogenesis of salivary glands and the face.
Yamada’s work on cell-matrix interactions sheds light on how cells interact naturally with the ECM and lays the groundwork for developing ideal microenvironments for tissue engineering.
From the 2012 Research Festival’s session “Matrix Biology and Matrix Remodeling,” chaired by Keir Neuman (NHLBI) and held on October 10, 2012.
Mysteries of the Microbiome
There are trillions of microbes on or in the human body—10 times as many organisms as the number of human cells—and most of them are beneficial to us. As part of the Human Microbiome Project (HMP), which NIH launched in 2007, researchers at universities, scientific institutions, and NIH are characterizing the microbes found in different regions of the body, including the nose, mouth, skin, digestive tract, and vagina. A few of the NIH intramural researchers gathered at the 2012 Research Festival to talk about their work.
Christian Abnet, an investigator in the National Cancer Institute’s (NCI’s) Division of Epidemiology and Genetics, is characterizing the upper gastrointestinal tract’s microbiome. In a large study—based on a population in Linxian, China, that is at high risk for developing esophageal cancer—he is investigating the roles of poor oral health and bacteria in the development of cancers of the esophagus and stomach. For example, Helicobacter pylori (HP) is known to increase the risk of esophageal and gastric cancers. In a nine-year follow-up study, Abnet’s team has observed that a less diverse oral microbiome is associated with the risk of death. Future studies will attempt to replicate these results in other populations and will examine which features of microbial diversity are associated with cancer risk.
Laurel Lagenaur, a collaborator scientist and guest molecular biologist in NCI’s Vaccine Branch, discussed her research on biotherapeutic approaches to preventing human immunodeficiency virus (HIV) infection. In animal models, she used Lactobacillus—a genus of commensal bacteria, most of whose members convert lactose and other sugars to lactic acid and are found in the vagina and gastrointestinal tract—to promote the production of the HIV inhibitor protein cyanovirin-N (CV-N). Lactobacilli may protect against bacterial vaginosis, and CV-N prevents HIV entry. Successful test results in female macaques showed that colonizing the vagina with Lactobacillus bacteria that produced CV-N protected against a lab-made strain of HIV engineered to infect monkeys. Lagenaur’s results may be translated for human clinical trials. Her aim is to facilitate disease prevention in a safe, low-cost manner in high-disease locales in the developing world.
Others who presented their microbiome work at the festival included Shruti Naik (NIAID), Katia Cargia-Crespo (NIAID), and Noriho Iida (NCI and FARE award winner), as well as several who presented posters. The symposium even spurred an initiative to hold meetings to further discuss microbiome research and explore collaborative opportunities. For more information on the HMP, visit https://commonfund.nih.gov/hmp or https://irp.nih.gov/catalyst/v20i5/research-briefs.
From the 2012 Research Festival’s session “Microbiome Research at the NIH: From Disease to Therapeutics,” chaired by Rashmi Sinha (NCI) and Leigh Greathouse (NCI); sponsored by the Microbiome Working Group and held on October 10, 2012.
This page was last updated on Friday, April 29, 2022
