Intramural Detectives
Investigate Klebsiella Mystery
“Microorganisms can live in all sorts of extreme environments,” Chief of Clinical Care Medicine Henry Masur told an audience at a Clinical Center (CC) Grand Rounds in September. “They can live without oxygen . . . in boiling water . . . on ice, and they can live with all sorts of antibiotic pressure.”
That ability to resist antibiotics can transform even harmless bacteria into dangerous pathogens that can sicken hospitalized patients, especially those with already weakened immune systems. In the summer of 2011, a strain of Klebsiella pneumoniae that was resistant to the powerful antibiotic carbapenem began working its way through some of the CC’s most gravely ill patients. Even infection-control procedures failed to stop the spread before seven patients died.
“We were deeply saddened by their deaths,” said Clinical Center Director John Gallin. “We were determined to stop this infection in its tracks and do everything possible to protect our patients.”
So the Clinical Center did what no ordinary hospital could. It marshaled the forces of NIH’s intramural program: Intramural Sequencing Center scientists and technicians, CC microbiologists and epidemiologists, and National Human Genome Research Institute researchers pitched in to help.
The story was first told in the August 2012 issue of Science and Translational Medicine, then retold—sometimes incorrectly—by dozens of national media outlets. Finally, at the September grand rounds, CC epidemiologist Tara Palmore and NHGRI senior investigator Julie Segre joined Masur (who was not one of the authors on the paper) to describe how skillful sleuthing, advanced genomic technologies, epidemiological practices, and surveillance enabled the NIH team to hunt down and contain an elusive bacteria.

MAGGIE BARTLETT, NHGRI
Left to right, Evan Snitkin, Tara Palmore, and Julie Segre (above) were part of the intramural team that investigated the cluster of Klebsiella pneumoniae cases among gravely ill, immune-compromised patients at the NIH Clinical Center.
It all began on June 13, 2011, when a woman (Patient 1) with a rare lung disease was transferred from a New York City hospital to the 243-bed CC hospital. She was known to be colonized with (a carrier of) a strain of carbapenem-resistant Klebsiella pneumoniae (KPC). The clinicians took every precaution to keep it from spreading. She was kept separate from the rest of the patients and placed in a private room; staff and visitors had to wear gloves and gowns when entering; and strict hand-washing protocols and other infection-control procedures were in place. She also spent two 24-hour periods, in isolation, in the intensive care unit (ICU).
Klebsiella is a normally harmless bacteria found in the intestines. It can, however, cause difficult-to treat-infections—such as pneumonia and bloodstream infections—in immunocompromised patients. Hospital-acquired infections—such as methicillin-resistant Staphylococcus aureus, Clostridium difficile, KPC, and others—complicate about five percent of hospital admissions across the country and kill nearly 100,000 people a year, according to the Centers for Disease Control.
When Patient 1 was released from the CC on July 15, it appeared as if the KPC had not spread. But three weeks later, another patient, who had never been exposed to Patient 1, tested positive for the organism. Eventually the CC’s first-ever cluster of KPC infections, which was largely ICU-based, involved 19 patients, seven of whom died from the infection. (The latest death was in September 2012; Patient 1 is still alive.)
It’s not uncommon for bacteria to lurk silently and pop up again weeks after their source is gone. Still, the doctors wondered whether the cases were linked to Patient 1 or whether there were other sources of KPC.
The usual methods of investigation would not be enough to solve the mystery. Standard molecular typing methods—such as pulsed-gel electrophoresis—could identify similar strains of Klebsiella but were unable to detect the small genetic differences that could provide clues about how the infection was spreading. Fortunately, NIH intramural researchers have access to powerful rapid-genetic-sequencing tools that can map entire genomes in a day (the process used to take years and be prohibitively expensive) and to sophisticated methods for analyzing the information.
Bioinformatics specialist Evan Snitkin, a postdoctoral fellow in Segre’s lab, developed an algorithm to reconstruct the outbreak transmission based on whole-genome sequencing and epidemiological data that traced the location of patients throughout their hospital stay.
By observing changes in the nucleotides as the bacteria reproduced, we “were able to track the evolution of this organism,” to other patients, said Segre. The KPC strains in all 19 patients could be genomically traced back to Patient 1.
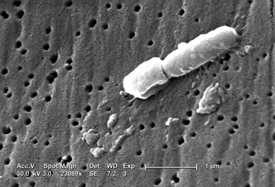
JANICE CARR, CDC
This scanning electron micrograph (above) reveals some of the ultrastructural morphologic features of a Klebsiella pneumoniae bacterium.
The researchers also used extreme measures in their investigation. “We threw the kitchen sink at these hospital-acquired organisms,” quite literally removing and decontaminating six sink drains colonized with the bacteria, said Palmore. Although a ventilator remained contaminated after thorough cleaning, it was not used on patients afterwards and therefore not linked to spreading the infection.
NIH is the first organization to use real-time, whole-genome sequencing to track an infection of this sort. Eventually, as the technology advances and its costs decline, hospitals will be able to “apply these novel technologies to track hospital transmission routes and find specific opportunities for intervention,” said Segre.
In the case of the CC, the findings reinforced the need to adhere to stringent infection-control precautions. The CC even hired monitors to make sure everyone was following all the protocols.
“The cooperation of the entire Clinical Center staff, and our patients and their families to stop transmission has been extraordinary,” said Gallin. “It’s a model for other hospitals.”
To read the journal article go to http://www.sciencemedicine.org/content/4/148/148ra116.full. (Sci Transl Med 4:148ra116, 2012).
To view the September 5, 2012, CC Grand Rounds, visit https://videocast.nih.gov/launch.asp?17542.
This page was last updated on Friday, April 29, 2022
