NIH Research Festival Highlights
Dynamic Protein Assemblies
Welcome to the gap . . . in how we visualize and understand biological structures, that is. Classical methods of determining biological structures have given us beautiful data about the very small (small, well-ordered protein and RNA molecules) and the very large (whole cells or organelle structures). But many biological mysteries surround the large, dynamic protein assemblies that are responsible for complex biological processes and yet cannot be seen by the classical methods of structure determination.
Historically, structural biology has come to mean X-ray crystallography or nuclear magnetic resonance (NMR) spectroscopy or, more recently, cryo-electron microscopy (cryo-EM) technology, said Sriram Subramaniam (NCI), a co-chair of the “Dynamic Protein Assemblies: Large and Small” session. However biological structures featured in this symposium were beyond the traditional applications of these methods.
Many of the session’s six presenters either combine multiple methods or use traditional methods in new and unique ways to examine the heterogeneous, dynamic molecular complexes that lie at the heart of many biological phenomena.
“This integration of different modalities allows us to look at biology at multiple scales, to go seamlessly from the atom-by-atom scale to the cellular scale and beyond,” said James Hurley (NIDDK), the other session co-chair.
The opening presentation by FARE winner Nicholas Noinaj (NIDDK) was a perfect example of this idea of integrating different modalities. Noinaj used X-ray crystallography, small-angle X-ray scattering (SAXS), and EM to look at the structure of the transferrin receptor in Neisseria bacteria. Neisseria is associated with meningitis and gonorrhea. The transferrin receptor participates in stealing iron from host transferrin, a glycoprotein that binds iron. Deciphering this structure, Noinaj said, “represents a significant advancement in our knowledge of iron import by bacterial pathogens” and offers new data that will be useful for vaccine and drug development.
The next presentation, by Adriaan Bax (NIDDK), addressed the well-studied influenza hemagglutinin envelope glycoprotein. Using a DNA-based liquid crystal medium for NMR spectroscopy, Bax found that the two helices of the glycoprotein’s membrane anchor form a tight hairpin structure within the membrane, stabilized by hydrogen bonds between the helices. Bax also found that the entire hairpin structure can rotate within the membrane. “The remarkable and highly unusual structure of [the envelope glycoprotein] fusion domain suggests a distinct function in the fusion process,” said Bax. His findings are contrary to what’s been reported in the literature—that the structure is a passive membrane anchor. “Don’t take everything you read in the literature as a fact!”
Hurley also presented data from his own group and collaborators, including Gerhard Hummer (NIDDK), on the endosomal-sorting complex required for transport (ESCRT). The ESCRTs, Hurley explained, are too flexible to crystallize, too small for EM, and too big for NMR. Using SAXS, double-electron electron-resonance spectroscopy, electron–paramagnetic resonance spectroscopy, and single-molecule fluorescence resonance energy transfer, Hurley and colleagues determined both open and closed positions for the ESCRT protein complex. From this collection of structures, Hurley described a banana shape, whereby the ESCRTs force membrane curvature and induce membrane scission. The goal, Hurley said, is to “ultimately create a step-by-step molecular movie of how the budding scission and cargo-transport process works.”
Although the structures of many proteins have been described, Yun-Xing Wang (NCI) pointed out that technical challenges have prevented scientists from determining many structures for messenger RNAs (mRNAs) despite their biological abundance and importance. Using SAXS, Wang presented a global topological structure of a regulatory protein—the Rev Response Element (RRE)—from the mRNA of the human immunodeficiency virus (HIV). He found that in three dimensions, two protein-binding sites were brought into close proximity. Determining the RRE topology structure resolves the long-running mystery of how HIV virus selects its own mRNA, not the host RNA, for packaging.
The question addressed by Wei Yang (NIDDK) also involved protein–nucleic acid interactions, especially the recognition of DNA damage by the MutS protein complex. “There are unlimited varieties of DNA lesions and different pathways to repair damaged DNA,” said Yang. She found that the increased flexibility of damaged DNA, rather than the mismatch itself, is recognized by the MutS protein, and “the energy derived from ATP [adenosine-5’-triphosphate] hydrolysis can be used to increase the accuracy of DNA lesion identification.”
Subramaniam gave the session’s final presentation on the use of cryo-EM to obtain high-resolution images of large protein structures including bacterial chemotaxis receptor clusters and the GroEL chaperone complex. “The work I presented is very much on our road map of looking at large, heterogeneous, complex assemblies,” he said. He and his colleagues are “not satisfied with describing them as blobs.”
The field of structural biology is changing. “A seamless integration across scales and fields is where it’s going,” said Hurley. “In the future, we may not really talk about structural biology. It may be viewed as the highest resolution and the most quantitative dimension of molecular and cell biology.”
This session was co-chaired by Subramaniam and Hurley.
Capturing the Complexity of the Transcriptome
NHGRI
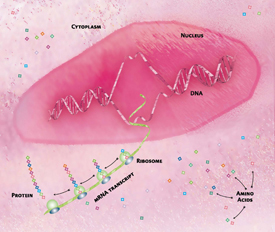
To make a protein, a gene’s DNA sequence is transcribed into messenger RNA (mRNA). This molecule, referred to as an mRNA transcript, leaves the nucleus and enters the cytoplasm. Ribosomes then read the transcript and use the information to assemble amino acids into proteins. The entire RNA sequence of an organism is called the transcriptome.
If you thought that figuring out the genome—the entire DNA sequence of an organism—was complicated, try figuring out the transcriptome—the entire RNA sequence.
The transcriptome represents a very small percentage of the genome—less than five percent of the human genome—that is transcribed into RNA molecules. It includes messenger RNA (mRNA), which is translated into proteins as well as other types of molecules, which may influence cell structure and regulate genes. Because each gene may produce many types of RNA molecules, the transcriptome is more complex than the genome that encodes it.
Scientists compare transcriptomes of different types of cells to gain a deeper understanding of how each cell type functions and how changes in the normal level of gene activity may reflect or contribute to disease. By aligning the transcriptome of each cell type to the genome, it is possible to generate a comprehensive, genome-wide picture of which genes are active in which cells.
At the “Notes from the RNA-Seq Revolution: Deep Sequencing Transcribed RNA in Health and Disease” session, scientists from several institutes shared strategies for using a technique that is better than microarray profiling and other technologies at capturing the complexity of transcriptomes. RNA sequencing (RNA-Seq) uses high-throughput sequencing technologies to provide precise estimates of transcript abundance; catalogue all species of transcripts (mRNAs, noncoding RNAs, and small RNAs); detect novel, low-abundance transcripts; determine the transcriptional structure of genes; and measure allele-specific expression.
Senior investigator Mark Cookson (NIA) is researching genes involved in RNA editing—a process in which an RNA molecule is altered through a chemical change—within the brain. He is particularly interested in the action of adenosine deaminase on the RNA (ADAR) gene family. During brain development, the ADAR-1 expression level increases. Using RNA-Seq, Cookson demonstrated that the expression change is due to RNA editing and not transcriptional regulation.
Andrew Oler (NIAID), a high-throughput sequencing bioinformatics specialist, is using RNA-Seq to analyze human and mouse platelet transcriptomes as well as identify platelet-specific genes and their major and minor isoforms. When a tear occurs in a blood vessel wall, the protease thrombin binds and cleaves the amino-terminus of protease-activated receptor 1 (PAR-1) to trigger the activation of platelets, which plug the tear. He showed that during platelet activation, PAR-1 expression is low in mice but high in humans and PAR-3 is increased in mice but low in humans.
RNA-Seq is even being used to help identify genes that contribute to the risk for bipolar, or manic-depressive, disorder. Senior investigator Francis McMahon (NIMH) and colleagues at NIMH are using RNA-Seq to analyze the RNA within the prefrontal cortex of people with and without bipolar disorder. In a small study that involved 10 subjects, they identified 97 differentially expressed transcripts involved in various pathways including cell-cell signaling, ion transport, and synaptic and nerve impulses.
Postdoctoral researcher and FARE winner Shurjo Sen (NHGRI) is trying to improve the RNA-Seq protocol while studying atherosclerosis in cardiovascular disease. He recommended certain equipment to achieve clean, fast, and reproducible RNA-Seq results and better data yield. In his atherosclerosis research, Sen examined cardiac computed-tomography scans to detect calcium buildup in the coronary artery and then compared high- versus low-calcification groups. He observed an increase in categories for cell–surface receptor signaling pathways and regulation of cell adhesion in the high-calcification group.
A hurdle for using RNA-Seq technology is the enormous amount of raw data generated for each sample. Bioinformatics expert Nirmala Akula (NIMH) is dealing with the challenges of processing, analyzing, and storing RNA-Seq data. Akula maps the data to a reference genome, focuses on the quality of base-pair readings to improve the percentage of sequences that are mapped,and uses computer software to tally the number of mappable reads.
The researchers expect that once they overcome the barriers to widespread use of RNA-Seq—higher cost, high data-storage requirements, and the absence of a gold standard for analysis—this technique will become the predominant tool for transcriptome analysis.
This session was co-chaired by McMahon and Cookson.
Bringing Stem Cells to the Clinic
Induced pluripotent stem cells (iPSCs) hold tremendous potential in many disciplines of biomedical research, both in the laboratory and in the clinic. Several of the NIH researchers who are exploring that potential described their work in a symposium entitled “iPSC Cells for Screening and Therapy.”
Kapil Bharti (NINDS) makes retinal pigment epithelium (RPE) from iPSCs in order to understand the mechanisms underlying macular degeneration and other eye diseases. At first, the iPSCs he created could not be efficiently differentiated into RPE cells and the generated cells were not fully authenticated. Once he used growth factors critical to the RPE development pathway, however, he was able to generate stable and authentic RPEs. Bharti stressed the importance of using reliable protocols to ensure that the newly derived cells duplicated their counterparts in the human body. Establishing this foundation allows RPEs and other iPSCs to be used in high-throughput screens and therapeutic development.
Indeed, iPSCs may be the foundation for new pharmaceutical therapies. For example, Heiner Westphal (NICHD) is collaborating on research with Forbes Porter (NICHD) that they hope will lead to the development of a drug that would improve cholesterol synthesis in patients with a developmental disorder known as Smith-Lemli-Opitz syndrome (SLOS). SLOS is caused by a mutation in a gene critical to cholesterol synthesis; patients have low cholesterol concentrations, which are thought to disrupt a variety of cells including neurons. Westphal’s group used iPSCs from SLOS patients to generate various neuronal cell types and developed an assay to measure the percentage of cholesterol precursors present.
Another type of stem cell therapy involves isolating and modifying a patient’s own cells to express a needed protein and then transplanting the modified cells back into the patient. FARE recipient Colin Sweeney (NIAID) presented research demonstrating how this approach could be used to treat the genetic disorder chronic granulomatous disease, which leads to long-term and repeated infections. In this disorder, a defect in the production of reactive oxygen species (ROS)—chemically reactive molecules containing oxygen—prevents immune cells from digesting engulfed bacteria and other infectious particles.
Sweeney is studying a form of the disorder caused by mutations in the membrane protein gp91. He generated iPSCs from patient cells and then used directed gene transfer to integrate the gp91 gene into the genome of the iPSCs. The resulting cells were differentiated into neutrophils and found to have ROS activity close to that of normal control subjects. Although Sweeney considers his work a successful proof of principle, he is trying to develop a better method whereby the modified cells could be more easily transplanted into patients.
The new Center for Regenerative Medicine focuses on clinically applicable and feasible iPSC research, explained session co-chair John O’Shea (NIAMS). He was the acting director for the center until Mahendra Rao (NIAMS) was appointed in the summer of 2011. O’Shea encouraged researchers to consider how they establish their cells, use them for clinical applications, and make them ready for ultimate delivery to a patient.
This session was co-chaired by Rao and O’Shea.
Immune–Targeted Therapies
One of the most powerful weapons in a clinician’s disease-fighting arsenal is the patient’s own immune system. The focus of the “Advances in Immune-Targeted Therapies” symposium was on therapies that use the immune system to fight or prevent diseases, even those outside the traditional viral and bacterial vaccine targets. The presenters showcased vaccine- and immunology-based treatments for several diseases, including vector-carried pathogens, cancers, and autoimmune disorders. They focused on recent advances in creating effective immune-targeted therapies and the common challenges for clinicians using these treatments.
“There has been a lot of interest [in immune-targeted therapies] because there has been an explosion of possibilities to treat different diseases using immunology-based technologies,” said Liliana Guedez (NCI), an organizer and co-chair of the session. As the first clinical trials in this field begin to produce data, the differences between animal models and humans have become increasingly apparent.
The six speakers covered diverse diseases, but each of the therapies being studied shared a common theme: With proper training, the immune system can affect remarkable, and sometimes long-lasting disease responses and recoveries in patients, even those who have failed to respond to previous therapies.
The first talk of the session was by session co-chair Shaden Kamhawi (NIAID), who presented research on vaccine development for the vector-borne pathogen Leishmania. “Our group led by Jesus Valenzuela really does believe that [the vectors] are a neglected component of these neglected diseases,” said Kamhawi. The vectors “are an intact source that could be very significant towards controlling some of these afflictions.” Kamhawi’s research explored the use of a vaccine directed against a molecule isolated from the saliva of one of the species of sand fly that carries the Leishmania parasite. This experimental vaccine induced protection against the Leishmania organism in mouse models.
The next talk, by Nicholas Restifo (NCI), addressed the use of adoptive cell transfer to treat late-stage metastatic cancers. In this method, “a patient’s own anti-tumor lymphocytes are removed from their body, expanded and activated, and then given back to them,” Restifo said. This process is, in essence, encouraging and expanding the patient’s own immune system to combat the cancer. A 93-patient trial of this method was presented in the session, and it showed complete tumor regression after five years in 20 percent of patients, a far higher rate of durable, complete responses than any current FDA-approved treatments. This new research “makes us realize that the immune system is very powerful and is likely to be part of a curative tumor treatment” for metastatic cancer, concluded Restifo.
Ravi Madan (NCI) approached immune-targeted therapies for cancer from a different perspective. Rather than tune immune cells directly, Madan combined cancer vaccines with traditional therapies, such as the radiopharmaceutical agent Quadramet (samarium-153 lexidronam injection). Although traditional cytotoxic therapies produce rapid cancer regression, the remaining cancer cells immediately return to their original growth rate after the therapy is concluded. The vaccine treatment, however, slows the overall rate of growth of the tumor. The combination of these treatments, Madan suggested, may further improve survival over traditional or vaccine treatments alone.
Continuing on the cancer theme, the next talk, by Alan Wayne (NCI), focused on the treatment of acute lymphoblastic leukemia (ALL). “Despite the great success” in treating ALL, Wayne said, it is still “a leading cause of mortality from cancer in pediatrics.” Wayne presented research on the novel recombinant immunotoxins BL22 and HA22, cytotoxic molecules fused to an antibody to the B-cell marker CD22. The cytotoxic molecules directly the overexpanded CD22 positive leukemia cell population in ALL. In clinical studies, Wayne found positive responses in approximately two-thirds of patients, even those with multiply relapsed disease. There are ongoing studies with clinical protocols designed to improve efficacy and reduce toxicity.
Bibiana Bielekova (NINDS) presented compelling new data from a phase 2 clinical trial using the antibody-based therapy daclizumab, a treatment for multiple sclerosis (MS). Daclizumab binds the alpha-chain of interleukin-2 and is effective alone or in conjunction with interferon-beta. Bielekova detailed the multiple modes of action of this treatment on the immune system in MS patients.
The final presentation of the session, by FARE winner Joanna Fares (NCI) concerned a study of the tumor suppressor p15Ink4b. This molecule, she reported, is involved in the maintenance of macrophages and dendritic cells and thus in tumor immune surveillance.
A common theme among the talks was the challenges of translating bench research to the clinic. Equally striking, however, was the diversity of approaches that fit under the umbrella of immune-system-based therapy. “Every day,” said Guedez, “you see more and more and more ways to use immunology to design medicines for a lot of diseases.”
This session was sponsored by the NIH Translational Research Interest Group and co-chaired by Guedez and Kamhawi.
When Neural Circuits Go Awry
Neural circuits in the brain can go awry in many neurological disorders. At the Research Festival’s minisymposium entitled “Neural Systems Underlying Social Function in Normal and Pathological Conditions,” several NIH scientists talked about their attempts to gain insights into the neural systems underlying autism spectrum disorders, schizophrenia, and childhood psychopathy.
Researchers first discussed functional connectivity analyses and complex social processes such as intersubject synchronization during natural conversation and conscious awareness. FARE award winner Eun Young Kim (NIHCD) talked about developmental synaptic N-Methyl-D-aspartic acid (NMDA) receptor remodeling by Kv4.2 potassium channels in vivo. Explaining how the Kv4.2 knockout mouse showed altered NMDA receptor expression and decreased maturation, Kim described the brain’s different, developing connected areas as requiring transport vehicles on a road with “rules, stop signals, and limits—all on the lookout for cops and accidents.” She found that an excitation-inhibition imbalance and early-life seizures can alter neural plasticity and trigger disorders such as schizophrenia and autism.
Next, Nuria Abdulsabur (NIDCD) presented research on a functional magnetic resonance imaging (fMRI) study that looked at brain activity during natural conversations. Also likening conversation to “a busy two-way street,” she explained that the routes of thought transit “often merge in a complex intersection.” In this research, they performed fMRI scans on pairs of friends engaged in four-minute conversations on a variety of subjects designed to elicit preset moods and thought processes. Abdulsabur then analyzed the data to assess the brain scan differences generated by topic and mood. Most notably, gender differences were greatest during debate and adversarial social task situations.
It’s also important to understand the neural basis of the fMRI signal. Biyu He (NINDS) discussed how a physiological correlate of the fMRI—the slow cortical potential [polarizations of the electroencephalogram (EEG) or magnetic-field changes in the magnetoencephalogram that last from 300 milliseconds to several seconds]—may be involved with the emergence of consciousness. Her study measured fMRI signals and brain-field potentials in neurosurgical patients while they were awake as well as asleep. She also discussed studies that involved testing neuronal activity in altered states of consciousness in both vertebrates and invertebrates.
Next, researchers presented findings on clinical disorders and the breakdown of neural connectivity. Stephen Gotts (NIMH) described a new study in which he used a novel whole-brain connectivity approach in fMRI to measure the covariation of brain activity in different parts of the brain. Normally, social behaviors involve an interaction between the limbic-related brain structures (amygdala, hippocampus, prefrontal cortex, and anterior temporal lobes) that are involved in the more affective and emotional aspects of social processing and the social brain regions that are involved in language, communication, and comprehension. Gotts found that for people with autism spectrum disorder (ASD) the limbic and nonlimbic areas are decoupled. He also found that this limbic decoupling is what predicts the severity of social impairments in the ASD subjects.
James Blair (NIMH) continued with a presentation on the neurobiology of conduct disorder (CD) with callous and unemotional traits. He discussed the difficulty with the current diagnosis of CD, which encompasses some individuals whose aggression relates to emotional volatility and mood and anxiety disorders (bipolar disorder, depression, post-traumatic stress disorder, pervasive developmental disorder) and other individuals with callous-unemotional traits whose reduced emotional responding protects them from developing mood and anxiety disorders.
He presented data showing that this second group shows reduced emotional response—and reduced activity in brain areas that allow these responses—to the distress of others (their fear, sadness, and pain). He also showed that this group faces significant challenges in learning reward-and-punishment information and in using this information to make good decisions. Brain areas—such as the amydala, caudate nucleus, and orbital frontal cortex—crucial for learning about, and using, this information show considerable disruption in youth with CD and callous-unemotional traits. He explained how these difficulties lead to their reduced guilt and empathy and increased risk for aggression.
Finally, Bruno Averbeck (NIMH) talked about his research on emotion perception and oxytocin in schizophrenia. People with schizophrenia are known to have difficulty recognizing emotions. They tend to misattribute neutral expressions as negative expressions, overattribute disgusted expressions, and underattribute happy expressions. Recent studies have shown that the neuropeptide oxytocin can have beneficial effects on social behaviors. Averbeck conducted an experiment in which he gave people with schizophrenia an emotion-recognition task: looking at pictures of faces showing emotions—anger, disgust, happiness, fear, sadness, and surprise—and trying to identify the dominant mood of each. Patients with schizophrenia who were given oxytocin fared better on correctly matching five out of six of the emotions (except for surprise) than did people given a placebo. Averbeck’s findings offer the potential of integrating oxytocin treatment with psychological therapy.
This session was co-chaired by Alex Martin (NIMH) and Averbeck.
This page was last updated on Monday, May 2, 2022
