Systems Biology as Defined by NIH
An Intellectual Resource for Integrative Biology
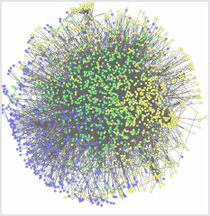
Seth Berger, Mount Sinai School of Medicine
It used to be as simple as “the knee bone connected to the thigh bone.” Now scientists use systems biology approaches to understand the big picture of how all the pieces interact in an organism. The above illustration depicts an interactome, the whole set of molecular interactions in cells. The interactome is considered an essential systems biology resource.
Ask five different astrophysicists to define a black hole, the saying goes, and you’ll get five different answers. But ask five biomedical researchers to define systems biology, and you’ll get 10 different answers . . . or maybe more.
Systems biology is an approach in biomedical research to understanding the larger picture—be it at the level of the organism, tissue, or cell—by putting its pieces together. It’s in stark contrast to decades of reductionist biology, which involves taking the pieces apart.
Yet with its complicated flow charts that can (in the words of T.S. Eliot) “follow like a tedious argument of insidious intent,” systems biology has scared away more than a few researchers. Still others fail to fully appreciate its usefulness because it lacks a concise, unified definition.
“There [are] an endless number of definitions,” said Ron Germain, chief of NIAID’s new Laboratory of Systems Biology, NIH’s first organized foray into systems biology, which has been nearly a decade in the making. “It’s even worse than the elephant,” that infamous elephant that stymies the attempts of blind men to describe it because each feels just one part.
“Some people think of it as bioinformatics, taking an enormous amount of information and processing it,” Germain said. “The other school of thought thinks of it as computational biology, computing on how the systems work. You need both of these parts.”
The new NIAID lab reflects an intellectual journey that Germain and some of his NIH colleagues embarked upon as the Human Genome Project was nearing completion.

If the NIAID Systems Biology Laboratory were a symphony orchestra, lab chief Ron Germain would be its conductor-musician. As conductor, Germain provides the necessary structure for tempo and harmony. As musician, he provides the immunology base, essentially the study of macrophages. He has recruited "orchestra" members for the lab who have the skills to work on their own but are also able to work together in the name of systems biology.
At that time, circa 2001, biology was rich in genomic data; proteomics had come of age; and immunologists had identified many cellular and even molecular components of the immune system. Yet predicting immune system behavior remained as elusive as ever.
Whether a systems biology lab could tease out answers was far from clear. But despite the risk, NIAID Director Anthony Fauci and Scientific Director Kathy Zoon committed a steady stream of resources. Together with Germain, they hoped for, and threw their energy into, a new approach to understanding the immune system that would better embrace experimental and computational techniques to explore connections in all their intricate glory.
The new lab, formed in early 2011 from the Program in Systems Immunology and Infectious Disease Modeling (PSIIM), comprises Martin Meier-Schellersheim, head of the Computational Biology Unit; Iain Fraser, head of the Signaling Systems Unit; Aleksandra Nita-Lazar, head of the Cellular Networks Proteomics Unit; John Tsang, head of the Systems Genomics and Bioinformatics Unit; and Germain, chief of NIAID’s Lymphocyte Biology Section, providing the immunology base to this operation.
Independently, the unit heads interact with labs at NIH and beyond to establish and incorporate systems biology methods. In true team spirit, they work together to attack the most basic elements of immunology such as a response to an infection or vaccination.
Ironically, to best understand this new lab, we should take a reductionist approach to defining its parts. The system, it seems, is more than the sum of its parts.
Start with Computational Modeling
Sophisticated computational models and simulations represent integral parts of systems biology. In immunology, they are needed to understand the complex biochemical networks that regulate the interactions among the immune system’s cells and between these cells and infectious organisms.
Enter Martin Meier-Schellersheim, a physicist by training. He was the first to join NIAID’s venture in 2001, even before the launch of PSIIM. He has been most successful in empowering non–computational biologists to do computational biology. Indeed, he has helped foster the very team concept that underlies the new lab; his software brings advanced computational capacity to a broad range of biologists.
This willing involvement of biologists is paramount because models need solid experimental data as input and as a reference to ensure reality checks. Otherwise the biological models are likely to be oversimplified either for lack of data or because their development suffers from poor communication between experimentalists and theorists.
Meier-Schellersheim’s primary software tool, called Simmune, facilitates the construction and simulation of realistic multiscale biological processes. He is also involved in the ongoing development of a systems biology markup language, SBML3, that can encode advanced models of cellular signaling pathways.
Add Some Cell Biology
Iain Fraser, a biochemist and molecular biologist interested in the mechanisms of cell signaling, arrived at NIH in 2008. As the lead high-throughput member of the lab, he has several powerful tools on hand to generate key data sets. These data sets ultimately feed into Meier-Schellersheim’s software to produce quantitative models.
Fraser’s tools include in-house genome-wide RNAi screens to characterize signaling network relationships in hematopoietic cells. Such screens are beginning to identify key components in innate immune pathogen-sensing networks. He interacts closely with the NIH-wide RNAi screening group at the NIH Chemical Genomics Center and also with the RNAi Global consortium.
Fraser said immune-system signaling networks can be unraveled by using proper systematic approaches to interpret complex data sets. He offers the example of Toll-like receptors (TLRs), which trigger an intricate cellular response that activates multiple intracellular signaling pathways.
Excessive activation can lead to chronic inflammatory disorders; insufficient activation can render the host susceptible to infection. Unbiased screening approaches can help identify the components that allow the immune system to maintain a homeostatic balance in the face of microbial challenges.
One of Fraser’s early successes, using a systems biology approach, was demonstrating how a single protein kinase can mediate the anti-inflammatory effects of cyclic adenosine monophosphate in its crosstalk with TLR4.
Fraser sums up his time at NIH as establishing “the screening infrastructure for dissecting the response of the macrophage to a broad range of pathogenic stimuli.”
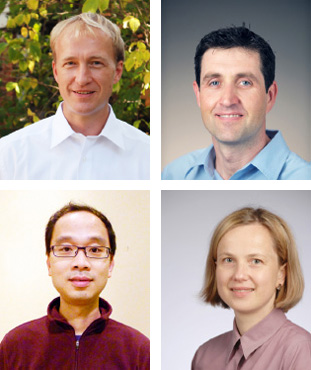
The “orchestra” members for NIAID’s new Systems Biology Laboratory include (clockwise from top left) Martin Meier-Schellersheim, head of the Computational Biology Unit; Iain Fraser, head of the Signaling Systems Unit; Aleksandra Nita-Lazar, head of the Cellular Networks Proteomics Unit; and John Tsang, head of the Systems Genomics and Bioinformatics Unit.
One Generous Serving of Proteomics
Aleksandra Nita-Lazar is developing new methods to obtain quantitative data that improve our understanding of cell biology and also funnel key information into model building. Her domain is the system-wide analysis of the proteome, which has fallen behind DNA analysis partly for want of the necessary tools.
The difference stems from the accommodating nature of DNA. DNA is easily recognized, replicable, and relatively stable, whereas the folded structure of proteins can’t be amplified. Yet protein studies are essential in developing useful models for many reasons, Nita-Lazar said. Such studies can reveal the molecular constituents of a cell; provide information about the biochemical state of the proteins; and determine catalytic rates and the association and disassociation rates for molecular pairs.
Nita-Lazar uses mass spectrometry to investigate protein phosphorylation, the process of binding with a phosphate group, one of the most common modes of protein-function regulation. She can use the same protocols that Fraser helped develop, and the same cell types, to determine which proteins are phosphorylated in response to a particular stimulus, when they are phosphorylated, and how those data fit into what is known about the transcriptional response.
Nita-Lazar’s group, with Fraser’s group, has been harvesting from these screens the key components required for the signal to flow through a pathway and also for the induction of the inflammatory cytokine messenger RNAs that arise. “This kind of approach used to be dismissed as a fishing expedition,” said Nita-Lazar. The goal is not to catch that one big tuna, however nice that would be, but rather to see the whole school of fish, the entire ecosystem.
Mix Well with Genomics
The enormous amount of data being collected requires processing and analysis—computational tools plus genetics and genomics “to build things from the top down,” Germain said.
Enter John Tsang, the most recent member of Germain’s lab and the element that transformed the PSIIM into a full-fledged systems biology lab.
On the genomics side, Tsang collects and analyzes data on gene expression, miRNAs, epigenetic modifications, and commensal microbes, and he conducts experiments to connect signaling to gene expression. On the bioinformatics side, he develops and applies statistical tools for large and diverse data sets, such as data from microarrays and high-throughput screenings, with an eye toward network models that involve genes, proteins, miRNAs, and epigenetic states.
Tsang also heads bioinformatics at the trans-NIH Center for Human Immunology (CHI), using similar integrative genomics approaches to study the human immune system, such as immune reactions to the flu vaccine in patients.
A core theme for building network models is capitalizing on systematic perturbations and -omics technologies to measure genome-wide responses. From the TLR stimulations that Fraser studies to vaccinations and natural genetic variations in humans, “all are valuable perturbations to help us figure out the wiring and function of the underlying system,” said Tsang.
Oh, Right, Immunology Too
“So, what am I doing in all of this aside from raising money and pontificating?” Germain joked.
If the NIAID Systems Biology Laboratory could be considered a symphony orchestra, Germain would be its conductor-musician. As conductor, he provides the necessary structure for tempo and harmony. As the musician, he provides the immunology base, essentially the study of macrophages.
Germain has seen systems biology labs in which collaborations are more opportunistic than routine, the shortsighted result of building a building, adding smart people, and hoping it all works out. His strategy instead has been to recruit individuals with the necessary skill sets to work on their own but also to work together in the name of systems biology.
“We all have slightly different interests, but there is enough overlap between those interests for us to develop those core projects and for us to be invested in them,” said Fraser.
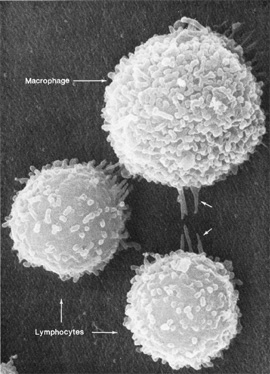
Don W. Fawcett, Emma Shelton
Macrophages and lymphocytes, the two types of immune cells pictured above, interact with their surroundings in complicated ways. NIH researchers are using systems biology approaches to understand the totality of such interactions.
All Together Now
To understand the response to infection or vaccination at an integrated level, the lab is studying the intersection of innate and adaptive receptor-dependent pathways and their control of gene networks. The researchers have bottom-up projects to understand and model the signaling within specific cell types at a fine-grained level.
And they have a top-down approach that uses inferences from perturbation analyses to probe the large-scale structure of the interactions not only at the cellular level, but also at the tissue and even the organism level.
To accomplish this grand goal, Germain said, the lab works in digestible chunks, focusing on pathogen sensing in key innate cells, such as macrophages, and the intersection of signaling by antigen receptors, cytokines, and TLRs in determining whether B cells become memory cells or long-lived plasma cells.
This process is critical for vaccine development. At the top-down level, the lab uses host genetics and microbiota variation to explore how the immune system’s set point is determined for responses to infections and vaccines.
This early into the chase, the lab has not yet published results on these pursuits, although a paper is pending on the lab’s work with CHI and the flu.
Towards a Trans-NIH Approach
Germain hopes the Laboratory of Systems Biology will serve “as an intellectual resource for people who are thinking in the systems mode and have their hands on these technologies [and want] to see how they could be applied to their work.”
He named the lab the Laboratory of Systems Biology with no mention of immunology or host-pathogen interaction to designate its raison d’être. Inspired by NIAID’s efforts, NCI and NHLBI are actively recruiting researchers to establish systems biology programs. NHLBI has just named Keji Zhao, senior investigator, as director of its new Systems Biology Center.
Meanwhile, the trans-NIH effort for a Center for Systems Biology is not dead. A search for a very senior systems biologist to develop and lead the center came up dry, and now the budgetary stresses have put the search on hold. But most NIH researchers understand that purely reductionist approaches to biology are no longer enough to solve complex biological problems and that integrated approaches are needed. David Levens (NCI), Dan Camerini (NIDDK), and Alan Michelson (NHLBI), along with Germain, continue to lead efforts for this trans-NIH initiative.
NIAID’s Laboratory of Systems Biology is “a smaller model of what the larger enterprise could be,” Germain said. The new lab “is very good for the NIH. We are getting applicants from top universities who want to come to the lab as fellows.”
And the NIH intramural research program is well suited for systems biology, with a long-term perspective and a retrospective review process that doesn’t require grant writing.
Germain helped change the NIH tenure process, too, to be sure that team science, and not necessarily a steady stream of published papers, is recognized and rewarded.
“Nothing happens if you don’t put work into it,” he added.
Reporter’s note:
Ron Germain does have his own definition of systems biology that he’s sticking to: a scientific approach that combines the principles of engineering, mathematics, physics, and computer science with extensive experimental data to develop a quantitative as well as a deep conceptual understanding of biological phenomena, permitting prediction and accurate simulation of complex (emergent) biological behaviors.
For more information about the Laboratory of Systems Biology, visit http://www.niaid.nih.gov/labsandresources/labs/aboutlabs/lsb/Pages/default.aspx.
This page was last updated on Monday, May 2, 2022
