Machine Learning

CREDIT: ALEXLMX, THINKSTOCK.COM
Teaching Computers to Think
Enter Richard Leapman’s lab at the National Institute of Biomedical Imaging and Bioengineering (NIBIB) and you’ll find a serial block-face scanning electron microscope, the size of a lab freezer, busily slicing and scanning pancreatic tissue, blood platelets, or other biological matter. In 12 hours, there will be 20,000 images of 25-nanometer-thick slices of each sample.
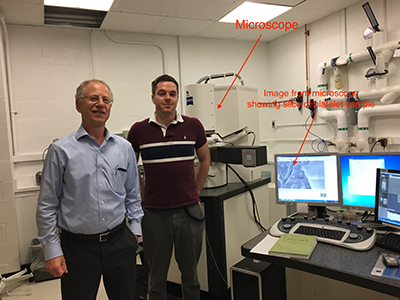
PHOTO BY SUSAN CHACKO, CIT
Richard Leapman (left) and Matt Guay (right) found that a machine-learning program could analyze the biological images produced by the block-face scanning electron microscope (behind them) much faster than a team of postbacs could.
Before the microscope arrived four years ago, Leapman had a team of postbacs manually delineating features of the objects in the images. Such work is labor-intensive, taking a hardworking postbac a few hours to trace the outline of a single platelet in an image set. Small wonder, given that a platelet that measures two to three micrometers across would be contained in about 80 image slices. It soon became clear that even the most zealous postbac would spend a lifetime working through the volume of data being produced by the microscope. When Matthew Guay joined Leapman’s lab as a postdoc data scientist, he suggested they use new developments in machine learning to speed up the process.
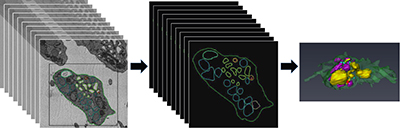
CREDIT: LABORATORY OF CELLULAR IMAGING AND MACROMOLECULAR BIOPHYSICS, NIBIB
A mouse blood platelet has been sliced into 80-some 25-nanometer-thick layers (left) by a serial block-face scanning electron microscope. It takes a person a full day to manually trace the organelles through all the slices of one platelet (center). On the right is the resulting 3-D model of the platelet’s subcellular organelles. With machine learning, computers can be taught to do the job in minutes. The sample was provided by Brian Storrie, University of Arkansas for Medical Sciences, Little Rock, Arkansas; data were analyzed by postbaccalaureate students Michael Tobin and Rohan Desai in Richard Leapman’s laboratory in NIBIB.
Machine learning is by no means new. It’s been around for decades. But thanks to big data and more-powerful computers, it has evolved into an amazing tool that has helped to advance—and often speed up—scientific research. In machine learning, computer systems automatically learn from experience without being explicitly programmed. A computer program analyzes data to look for patterns; determines the complex statistical structures that identify specific features; and then finds those same features in new data. For Leapman’s platelet images, a machine-learning program would start with the platelet structures that had been painstakingly outlined by the postbac team, learn the features that identify the structures, and then use computer algorithms to demarcate platelets and organelles in new sets of similar images at high speed.
A more recent outgrowth of machine learning is deep learning, which takes advantage of the availability of massive amounts of data and powerful computers to train large and complicated artificial neural networks. For supervised deep learning, each element of the training data is labeled according to whether or not it contains an object of interest; however, the researcher doesn’t specify to the computer which features uniquely identify that object. The computer learns to deduce the most-predictive features of the object from the dataset and then develops computational rules to identify labeled objects. This process is akin to pointing out cats to a small child, who will eventually figure out the features that identify a cat without needing to be told about ears or whiskers or tails.
Reliability, reproducibility, standardization, increased throughput, and decreased cost of data processing are among the benefits of machine learning. If larger datasets are available, deep learning can be used and is often more accurate than older machine-learning methods and less biased by human input.
Unsupervised machine or deep learning goes a step further: Objects or features are not labeled at all and the program searches for common characteristics to organize the data. This process offers the additional possibility of finding subtle patterns in large multivariate datasets such as genetic, imaging, and neurological data and biomarkers from patients.
Machine learning supports much of the technology around us: smartphone cameras recognizing faces; online translating and captioning; credit-card fraud detecting; personalized online marketing; and providing newsfeeds. In the NIH intramural program, machine-learning techniques are being used in several research areas: image processing; natural-language processing; genomics; drug discovery; and studies of disease prediction, detection, and progression.
Image processing
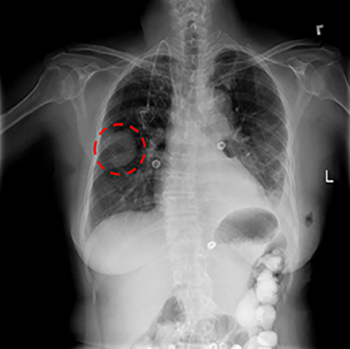
CREDIT: NIH CLINICAL CENTER
Ronald Summers’ group has been using machine learning and deep learning to improve the accuracy and efficiency of image analysis to enable earlier detection and treatment of diseases. His group recently released a curated set of 120,000 anonymized chest X-rays to the scientific community. Researchers around the world can use this publicly available dataset to develop automated techniques for disease detection and diagnosis. Shown: a chest X-ray showing a lung mass (circled in red).
Analyzing X-rays and other images. Ronald Summers’ group in the Clinical Center has been using machine learning for two decades so that computers could examine and analyze X-rays, computed tomography scans, and magnetic resonance images (MRIs). The goal is to improve the accuracy and efficiency of the image analysis to enable earlier detection and treatment of diseases. A few years ago, Summers and his group began using deep learning. “It has been the most accurate technique, a significant performance jump,” he said. His group recently released a curated set of 120,000 anonymized chest X-rays (CXRs) to the scientific community. Researchers around the world can use this publicly available dataset to develop automated techniques for disease detection and diagnosis.
Screening for diseases. At the National Library of Medicine’s (NLM’s) Lister Hill Center (LHC), Sameer Antani and George Thoma are developing deep-learning-based diagnostic methods that will improve disease testing, decrease its cost, and produce more accurate, reliable, and standardized results. Antani is using CXRs to automatically screen for tuberculosis (TB) and other pulmonary diseases, with a special focus on sub-Saharan Africa regions where medical facilities are limited and people have to travel long distances to hospitals. Ideally, sick people there need to be tested and treated close to home. A truck containing an X-ray generator and Antani’s software has been travelling around rural Kenya since 2015, allowing more Kenyans to get tested easily and treated early.

PHOTO BY SAMEER ANTANI, NLM
A truck containing an X-ray generator and Sameer Antani’s software (developed via machine-learning) has been traveling around rural Kenya, where medical resources are scarce and hospitals are often far away, since 2015, allowing more Kenyans to be screened for TB and other pulmonary diseases and treated early.
Thoma’s group is applying deep-learning models to screen for other diseases such as cardiovascular diseases, malaria, and the human papillomavirus (HPV). Sema Candemir is using deep learning—with CXR collections from international sources as well as from Ron Summers—to detect cardiomegaly (abnormal enlargement of the heart). Stefan Jaeger is applying a deep-learning algorithm to classify and count malaria parasites in blood-smear images faster and more accurately than humans can. Malaria affects millions worldwide, and inadequate diagnosis is one of the hurdles to overcome in reducing mortality. Jaeger developed a system on a smartphone attached to a microscope and is field testing it in Bangladesh and Thailand.
Screening for HPV is important because, if not prevented or caught early, it can develop into cervical cancer. Antani and his colleagues Zhiyun Xue and Rodney Long are working with Mark Schiffman, Nicolas Wentzensen, and their team at the National Cancer Institute (NCI) and collaborators at other organizations ( including Global Good, which is supported by Bill Gates and the invention expertise of Intellectual Ventures) to develop and validate an automated visual-evaluation (AVE) algorithm that will identify precancerous lesions during visual inspection of the cervix. This algorithm outperforms human interpretation and provides sensitive screening with minimal clinical training or cost. Installing AVE on camera phones or similar devices, combined with available low-cost treatments, could permit unprecedented dissemination of high-quality, point-of-care cervical screening, especially in developing countries where medical resources are scarce.
Thoma considers the results from applying deep learning to this automated screening “nothing short of a revolution.” LHC Scientific Director Clement McDonald concurs. McDonald was initially chary of the claims for this technique but now says that the results of deep learning have been truly impressive.
Analyzing complex brain-activation patterns. The National Institute of Mental Health’s (NIMH’s) functional MRI (fMRI) Core Facility, led by Peter Bandettini, supports structural MRI and fMRI for 70 clinical protocols including for schizophrenia, autism, anxiety disorders, multiple sclerosis, epilepsy, and stroke. Traditionally, fMRI studies compared scans from a patient group with those from a control group to identify specific differences related to a disease. Now, machine-learning and deep-learning methods can be used to analyze more complex and subtle brain-activation patterns as well as many other sources of information such as electroencephalograms.
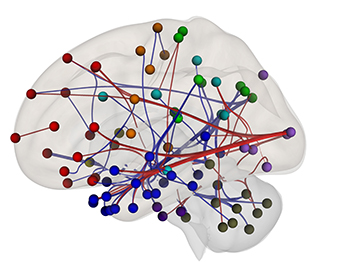
CREDIT: SECTION ON FUNCTIONAL IMAGING METHODS, NIMH
Neuroimagers are beginning to use patterns of “functional connectivity,” or correlations between certain brain regions’ fMRI activity, to predict a person’s performance on tests, their mental-health diagnosis, or their responses to certain kinds of treatment. These results could pave the way for a better understanding of reading difficulties, better interventions in schools, or even neurofeedback sessions that show struggling readers their own brain activity to help them improve their performance. Shown: fMRI scans were used to identify this “reading network” in which people with high functional connectivity in the red connections performed better on reading-comprehension questions than people with high functional connectivity in the blue connections.
“Deep learning is more of an adaptive model that allows scientists to find subtle connections without imposing a structured model on the data,” said Bandettini. “There’s a push to get more information from the research, have it actually translate to patients, and there’s also this overwhelming increase in data because of newer imaging techniques.”
Bandettini has recently started new collaborative teams within NIMH for data science and machine learning. The data-science team, headed by Adam Thomas, will share data across protocols so that intramural researchers can have access to well-curated image datasets and be able to mine data from public repositories. The machine-learning team, led by Francisco Pereira, will use various techniques—and train researchers to use them—including multivariate analyses to extract individual differences from fMRI data to predict which drugs may help treat particular conditions.
Screening for eye diseases. Machine learning is proving useful in detecting eye diseases such as age-related macular degeneration (AMD), the leading cause of incurable blindness worldwide in people over the age of 65. National Eye Institute (NEI) scientist Emily Chew, whose research focuses on AMD and other retinovascular diseases, is collaborating with Zhiyong Lu at NLM’s National Center for Biomedical Information (NCBI) to classify 60,000 retinal images and calculate risk factors for AMD. In their most recent work, the automated deep-learning technique was consistently more accurate than human ophthalmologists.
Chew is also working with Jongwoo Kim in Thoma’s group to develop deep-learning models that will improve screening for glaucoma. Drawing from thousands of images from NEI and international open-access sources, the model has learned to detect and classify features in specific regions of retinal images.
Natural Language Processing
In natural-language processing (NLP), researchers program computers to understand, interpret, and manipulate human language. Some of the ways that NLP is being used at NIH is for PubMed searches, dealing with medical questions that come from the public, and helping to improve the grant-application process.
Making PubMed searches better. Each day, some 2.5 million people use PubMed, NLM’s search engine that provides free access to over 28 million biomedical and life-sciences publications. Few are aware, however, of the machine-learning technology (developed by NCBI) behind it. PubMed traditionally sorts search results by date with the latest papers listed first. Zhiyong Lu points out that this method may not be ideal for satisfying various kinds of user information needs. In early 2017, his machine-learning work allowed PubMed users to search for relevance-based results by choosing the new sort option called “Best Match.”
Lu’s training dataset was developed by mining tens of thousands of past PubMed searches in an aggregated fashion. Dozens of features are used to rank the PubMed results, the most important being the past usage (how often the article was accessed), publication date, relevance score, and the type of article. Some papers might not contain the precise words in the query or even synonyms of those words but may still be relevant. Deep learning offers great promise here: It can identify related articles by analyzing words in context. And PubMed users, when responding to tests that present both the date-sorted and relevance-sorted (“Best Match”) results, have indicated that the relevance-based results are more useful to them.
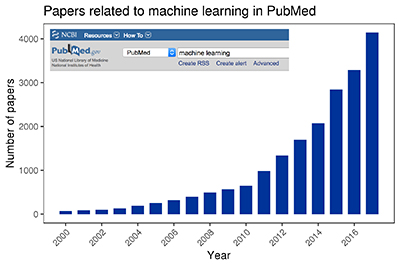
A PubMed search for machine learning shows the explosive growth of published research in this field since 2000.
Responding to medical questions. Streams of medical questions come in to NLM every day from people looking for advice and treatment options. An NLM team responds with suggestions pointing to accurate health information for patients and families at NLM’s MedlinePlus health-information website, but this process is both slow and expensive. Dina Demner-Fushman in Thoma’s group has developed prototype systems using NLP and deep learning to analyze incoming questions, extract question topics, and identify or generate precise answers.
Improving the grant-application process. The NIH Center for Scientific Review is the gateway for NIH grant applications and their review. To improve the grant-application process, Calvin Johnson’s group at the Center for Information Technology (CIT) has developed a tool that uses machine learning to analyze the text in the application and recommend an appropriate study section.
Disease Detection, Genomics, and Drug Discovery
Detecting diseases early. Parkinson disease (PD) is characterized by a wide variation in age of onset, duration until death, symptoms, and rate of progression. Until now, early detection and individualized predictions have been based on clinical observations. Andrew Singleton’s group at the National Institute on Aging is using deep learning to examine multidimensional data—clinical reports, imaging data, blood tests, genetic data, and results of neurological tests over time—from large cohorts of patients with PD.
Singleton points out that humans are very good at identifying patterns in 2-D or 3-D data, but only computers, using the latest techniques, can find subtle differences and patterns in 130 dimensions. The predictive models built by Singleton’s group allow for phenotypic clustering on a massive scale and indicates how patterns are related. The group recently used deep learning to identify the natural subtypes of PD as well as identify and predict the progression rates of each subtype. Singleton’s group is also collaborating with Sonja Scholz (National Institute of Neurological Disorders and Stroke) on a deep-learning project that analyzes genomic data to predict and differentiate among dementia syndromes.
Detecting gene-gene interactions. Joan Bailey-Wilson at the National Human Genome Research Institute (NHGRI) has been collaborating for decades with James Malley (retired from CIT and now a special volunteer at NHGRI) to develop open-source software and methods for statistical machine learning, with particular interest in detecting gene-gene interactions. They have been applying machine-learning methods to find genetic variants that increase the risk of complex traits such as childhood-onset schizophrenia and several hereditary cancers.
Accelerating drug discovery. Scientists at the National Center for Advancing Translational Sciences (NCATS) are collaborating with researchers at NIH and beyond to accelerate the discovery of therapeutic medications. In an effort led by NCATS Director Christopher P. Austin and Scientific Director Anton Simeonov, NCATS is developing and applying machine-learning models across a broad spectrum of drug-discovery projects including antiviral drugs, anti-infection medications, epigenetic targets and deubiquitinating enzymes, and medications for rare diseases.
NCATS researchers also use machine-learning approaches to guide the optimization of new drugs for adsorption and metabolism, select the best clinical candidate for efficient and safe clinical trials, and develop better toxicity-assessment methods. The models are available to other NIH institutes and centers and can be used, for example, to design RNA- and DNA-based nanoparticles with desired immunomodulatory activity as well as to predict toxicity, measure estrogen-receptor activity, and determine whether the stability of certain compounds depends on specific human cytochromes. Learn more about the 1,000-plus target-related models at https://predictor.ncats.io.
Challenges
Although deep learning holds huge potential in several biomedical areas, its success depends on getting large training datasets. Small training datasets can result in the program’s defining functions that do not generalize well to real-life data. Large annotated datasets can be difficult to obtain, however, so machine learning is a more practical approach for projects with small numbers (of patients, for example). In validating machine-learning methods, McDonald stresses how important it is to do a good statistical analysis of the datasets to avoid subtle biases.
Deep learning can be a black box. A scientist cannot easily tell why the algorithm comes up with a particular result. Singleton is sanguine: At the start “much technology appears [to be] a black box,” he said. “But if the results are reliable, that’s good enough to be getting on with [deep learning].”
The sensitivity of deep learning also means that the algorithms may not generalize well. A deep-learning program trained on, say, PubMed abstracts might not work well on full-text papers because the nature of the data is different.
In some cases, the algorithm can produce the “right result for the wrong reasons,” said Antani. His program once correctly identified a patient as having TB due to an abnormality in the X-ray that was actually not TB-related. To find the cause of such errors, Siva Rajaraman (NLM) is investigating the way in which the layers in a multilayer deep-learning structure process the input images.
What are the prospects for using deep learning in clinical diagnosis? Lu believes that physicians may have more trust in a traditional computer program with rules that can be understood and modified. An incorrect deep-learning prediction that is corrected by a physician might not change the next diagnosis: The algorithms learn from thousands of examples, so modifying a single sample may not change the way an algorithm works.
The heavy computational requirements mean that the training phase of deep learning takes longer than traditional methods. Once training has been completed, however, machine learning can be applied very rapidly.
Despite the challenges, machine learning is likely to produce impressive results in the future. “Previous methods limited the questions that could be asked,” said Bandettini. “But machine learning is exciting because it’s taking all of the data to the next level, [and] trying to make it usable for individual patients, as well as pulling even more subtle information out of it.”
Some Other IRP Researchers Using Machine Learning or Deep Learning
This is by no means a complete list.
Clinical Center
Scientific Director: John Gallin
- Daniel Joseph Mollura and Elizabeth C. Jones: Using machine learning to automatically and accurately evaluate imaging patterns related to various diseases.
Center for Information Technology
Scientific Director: Benes Trus
- Benes Trus: Collaborating with researchers in NICHD to develop an automated method to delineate neuroanatomical regions in zebrafish using in vivo imaging of gene-expression patterns from over 200 transgenic zebrafish lines; detection of variant types, prediction of target genes, and identification of subtypes of diffuse large B-cell lymphoma, in collaboration with NCI cancer genomic researchers.
- Matthew J. McAuliffe: Enhancing the Medical Image Processing Analysis and Visualization (MIPAV) application to rapidly build a machine-learning component integral to the MIPAV software that supports automated segmentation of prostate MRI images.
- Sinisa Pajevic: Applying machine-learning algorithms to study the relationships between gene-expression patterns and connectivity.
National Center for Biotechnology Information, National Library of Medicine
Scientific Directors: James Ostell
- Ivan Ovcharenko: Using deep learning to study DNA-sequence patterns in gene-regulatory elements, focusing on accurate identification of disease-causal mutations in enhancers and silencers of human genes.
- Anna Panchenko: Using machine learning to study cellular networks and how their perturbation can lead to diseases such as cancer.
- Teresa Przytycka: Developing deep-learning techniques for high-throughput analysis of short synthetic RNA and DNA molecules that bind to specific targets.
National Cancer Institute
Scientific Directors: Thomas Misteli, Glenn Merlino, and Bill Dahut
- Project Candle: NCI and the Department of Energy are collaborating on a project to develop machine-learning and deep-learning technologies to advance precision oncology.
- Peter Choyke: In collaboration with Ronald Summers’ group (Clinical Center), using a well-annotated prostate MRI dataset with associated histology data to develop a machine-learning-based diagnostic tool for prostate cancer. The tool has shown that pathologically significant tumors initially missed by radiologists provide a better definition of the true margins of visible tumors. The tool is currently undergoing testing in a worldwide multicenter trial.
- Yamini Dalal: Using machine learning to hunt for small molecules that can block aberrant chaperone-histone interactions that are cancer-specific in colon-tumor cells.
- Karl Thomas Ried: In research on disease prognostication and treatment response in rectal-cancer patients, used a machine-learning algorithm (trained on images from patients with rectal cancer) to build, train, and validate a predictor of pathological complete response.
- William Curtis Reinhold and Yves Pommier: Included a machine-learning approach to identify robust, cumulative predictors of drug response for 60 human cancer-cell lines.
- Xin Wei Wang: Using machine learning to identify biomarkers for early detection, diagnosis, prognosis, prediction, and treatment response of liver cancer.
National Eye Institute
Scientific Director: Sheldon Miller
- Tudor Constantin Badea: In research to study retinal circuit development and genetics, has been developing a machine-learning algorithm for the unsupervised detection and classification of retinal-ganglion-cell recordings from ex vivo retinas stimulated with a variety of visual stimuli.
- Bevil Conway: Using machine learning to classify images to gain insight into the neural mechanisms for object vision and to understand the extent to which color provides useful information about object identity.
National Human Genome Research Institute
Scientific Director: Daniel Kastner
- William Pavan: Using machine learning to evaluate the effect of variants in noncoding regions of the genome on the regulation of transcription.
- Francis S. Collins: In research involving the genetic analysis of type 2 diabetes in a Finnish population, developed and implemented machine-learning and deep-learning algorithms to predict DNA methylation values.
National Institute on Aging
Scientific Director: Luigi Ferrucci
- Jun Ding (in Schlessinger’s group): Created a program that uses machine-learning methods to measure physiological rates of aging of individuals.
- David Schlessinger (retired): Developed open-source tools for pattern recognition in biomedical imaging; used multiple machine-learning and deep-learning methods to analyze the aging of the eye microvasculature and the prognosis of macular degeneration and to assess individual rates of aging.
- Richard Spencer: Using machine learning to analyze cartilage degradation and osteoarthritis.
- Madhav Thambisetty: Implemented machine-learning approaches to understand preclinical pathogenesis and identify predictive biomarkers of Alzheimer disease.
- Bryan Traynor: Using machine learning to identify subgroups within people with amyotrophic lateral sclerosis (ALS) and to identify a model that allows the determination of age at onset of ALS and survival.
National Institute on Alcohol Abuse and Alcoholism
Scientific Director: George Kunos
- Abdolreza Reza Momenan and David Goldman: Using innovative approaches such as machine learning to assist in the neuroimaging of people with alcohol addiction to help with individualized treatment, measurement of treatment efficacy, and prediction of relapse.
National Institute of Allergy and Infectious Diseases
Scientific Director: Steve Holland
- Ron Germain: Combining machine learning and gene-expression signatures from diseased tissues for the discovery of blood biomarkers; using deep-learning methods to facilitate the analysis and interpretation of multiplex 2-D and 3-D confocal tissue-imaging datasets.
- Peter Kwong: Using machine learning to design prophylactic and therapeutic agents against HIV-1 and other human pathogens; developed programs that predict glycosylation patterns on viral antigens based on sequences and structures, protein solubility based on sequences, and antibody resistance based on HIV-1 isolates.
National Institute of Biomedical Imaging and Bioengineering
Scientific Director: Richard Leapman
- Hari Shroff: Uses deep learning to identify cells in light-microscopy images of roundworm (Caenorhabditis elegans) embryos in order to accelerate image analysis and eventually create a 4–D atlas of development in this animal model.
National Institute on Drug Abuse
Scientific Director: Antonello Bonci
- Kenzie L. Preston: Using phylogenetics and geospatial machine learning to map and predict HIV-transmission hotspots.
- Elliot Alan Stein: In research to determine the functional and structural brain circuits underlying smoking, has applied machine-learning techniques to fMRI data to predict neurological or neuropsychiatric disease states.
National Institute of Diabetes and Digestive and Kidney Diseases
Scientific Director: Michael Krause
Carson Chow: Using machine learning and deep learning for multiple projects—from fitting parameters of nonlinear models to experimental data, to inferring genetic variants in genome-wide association studies.
William A. Eaton: For a sickle-cell-disease research project, is developing a high-throughput screen based on nitrogen deoxygenation of sickle-trait cells and a robust machine-learning algorithm to determine the time at which an individual cell sickles.
Brian Clay Oliver: Developing machine-learning models to explain observed patterns of gene expression in Drosophila in a Drosophila functional-genomics research project.
Robert Star: Using deep learning to identify the filtering units of the kidney from biopsy-slide images and to improve the accuracy of automated biopsy assessment of disease severity and type.
National Institute of Mental Health
Scientific Director: Susan Amara
- Bruno Averbeck: Using machine-learning models to characterize behavior in learning tasks and to generate insight into the neural circuitry that underlies these behaviors.
- Karen F. Berman: In a project on neuroimaging of brain circuits and molecular mechanisms in normal cognition, used machine-learning algorithms for artifact screening and quality assessment of neuroimaging data, tasks generally requiring operator visual inspection.
- Sarah Hollingsworth Lisanby: Applying machine-learning approaches to data mining to support the discovery of common mechanisms across rapidly acting antidepressant interventions.
National Institute of Environmental Health Sciences
Scientific Director: Darryl Zeldin
- Clarice Weinberg and Leping Lu: Using machine-learning methods to study the effects of multiple mutations on the complex genetic associations associated with the birth defects cleft lip and cleft palate.
National Institute of Neurological Disorders and Stroke
Scientific Director: Alan Koretsky
- Bibiana Bielekova: In a comprehensive multimodal analysis of patients with neuroimmunological diseases, using machine learning to validate novel diagnostic classifiers, models, and clinical or imaging scales in independent validation cohorts.
- Leonardo Cohen: Applying supervised machine-learning techniques to brain activity recorded with magnetoencephalography to decode the temporospatial features of memory selection and maintenance during a working-memory task.
- Edward Giniger: In determining the role of the cluster of differentiation–5 kinase in neurodevelopment and neurodegeneration, used a combination of genome-wide expression profiling and machine-learning statistical methods to develop a novel metric for physiological aging.
- Kevin Lawrence Briggman: In mapping the synaptic connectivity and function of sensory neuronal circuits, used a machine-learning-based data-analysis pipeline to analyze a recently acquired mouse olfactory-bulb dataset.
Resources
- Deep Learning in Medical Imaging and Behavior SIG (Chair: Matt Guay): https://oir.nih.gov/sigs/deep-learning-biomedical-imaging-interest-group
- Data Science in Biomedicine SIG (Chairs: Ben Busby, Sean Davis, Lisa Federer, Yaffa Rubinstein): https://nihdatasciencesig.github.io
- Machine Learning Study/Discussion Group: (Chair: Lewis Geer): http://list.nih.gov/cgi-bin/wa.exe?A0=mlstudy
- Machine Learning LISTSERV: http://list.nih.gov/cgi-bin/wa.exe?A0=machinelearning
- NIH Biowulf: Intramural researchers have access to Biowulf, the in-house supercomputer with graphics-processing units that are particularly suitable for machine and deep learning: https://hpc.nih.gov
- NCATS 1,000-plus target-related models at https://predictor.ncats.io
- Pubmed: https://www.ncbi.nlm.nih.gov/pubmed/
- PubMed Labs, an experimental platform for features that may eventually migrate into PubMed: https://www.ncbi.nlm.nih.gov/labs/pubmed/
This page was last updated on Thursday, April 7, 2022
