The SIG Beat
NEWS FROM AND ABOUT THE SCIENTIFIC INTEREST GROUPS
NEW SIG: Metals in Biology Scientific Interest Group:
Metals are critical to a dizzying array of biological functions. Recent proteomic studies indicate that up to one-third of cellular proteins contain a bound metal cofactor, yet only a minority of these cofactors have been identified. Metal ions also bind and modulate the structure and function of RNA in an equally intriguing number of ways. By virtue of their variable coordination properties and redox behavior, metal ions often carry out unique functions in biology. However, metal ions such as copper and iron can be double-edged swords because, when unsequestered, they can generate toxic radical species that can damage proteins, nucleic acids, and lipids. Consequently, cellular systems devote a considerable amount of resources to the trafficking, chaperoning, and homeostasis of essential metal ions.
Malfunctions of one or more elements (pun intended!) of this metal-ion machinery cause many devastating human diseases such as hemochromatosis, Friedreich ataxia, Wilson disease, and Menkes disease. In addition, metal ions have proved valuable in medicine including serving as magnetic-resonance contrast-imaging reagents and anti-cancer therapeutics.
The Metals in Biology SIG unites several active intramural research groups whose interests span one or more aspects of the biology of metals. Although these groups are housed in different buildings and hosted by a variety of parent institutes, they have already formed thriving journal and data clubs that have been meeting informally for many years. The new SIG facilitates interactions among NIH researchers at the forefront of metallobiology and exposes trainees to a wider spectrum of problems, approaches, and techniques. The SIG plans to host one seminar every two months and continue the biweekly journal clubs. The inaugural seminar was given by Nigel Robinson from the University of Durham (Durham, United Kingdom) on June 11. For more information on the SIG, contact Anirban Banerjee (anirban.banerjee@nih.gov). To join the LISTSERV, go to https://list.nih.gov/cgi-bin/wa.exe?A0=METALSINBIOLOGY.
SGHD SIG: Sex Differences in Fruit Fly Sleep

CREDIT: IRYNA SHKRABALIUK, THINKSTOCK.COM
Fruit flies (Drosophila melanogaster) may hold the secret to helping scientists understand sleep. The little critters share many of the characteristics relevant to sleep in humans—they sleep mostly at night, stimulants can interfere with their sleep, and sleep deprivation can wreak havoc on their memory. In humans, sleep deprivation can lead to impairments in learning and memory, neurological diseases, cardiovascular problems, hypertension, and other disorders. Scientists can use fruit flies to study how genetic traits affect sleep, too. And some scientists like Susan Harbison are investigating something rarely studied: how sex differences may play a role in sleep.

PHOTO BY SARAH NEEDLE, NHLBI
NHLBI researcher Susan Harbison has found that male and female fruit flies have different sleeping habits.
Harbison, who joined NIH in 2012 and is a Stadtman tenure-track investigator in the National Heart, Lung, and Blood Institute, gave a presentation on her research at the May 22, 2018, gathering of the Sex and Gender in Health and Disease (SGHD) Scientific Interest Group (SIG).
But how do you measure sleep in a creature as tiny as a fly? You use an infrared-based Drosophila activity-monitoring system: Each fly is placed in a three-inch long glass tube; when it is awake and active, its movements disrupt an infrared beam; and the data can be used to decipher sleep.
Some of us require more sleep than others. To investigate this trait in flies, Harbison genetically engineered 19 long-sleeping and 20 short-sleeping lines. The long-sleeping flies slept for 17.8 hours in a 24-hour period, while the short-sleeping flies slept for only 3.3 hours. That’s a staggering 14.5 hours difference.
Harbison induced sleep deprivation by disturbing the flies mechanically via a custom-mounted vortexer. The device was programmed to deliver a shaking stimulus once a minute for 12 hours during the night. Harbison observed the flies’ rebound response (making up for lost sleep) the following morning. “Flies don’t have a job, so they can sleep in!” Harbison said during her talk. She found that short-sleepers lost less sleep (up to 412 minutes) than long-sleepers (up to 709 minutes) but the rebound response was similar (up to 260 extra minutes of sleep). However, there was a fair amount of variability among the lines, suggesting that the amount of rebound sleep is more dependent on genotype than sex. Preliminary data also showed that intermittent disruptions also affected sleep; short-sleeping males had a decreased arousal threshold (it takes less stimulus to awaken them) compared to females.
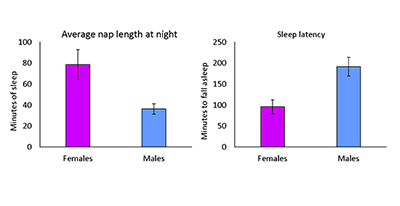
CREDIT: SUSAN HARBISON, NHLBI
Sex-specific differences in sleep. In this short-sleeping line, female flies have longer nightly naps, while male flies have longer sleep latencies; that is, they take longer to fall asleep once the lights go out.
As with humans, flies have a 24-hour circadian rhythm pattern. Using a rhythmicity index (the extent to which activity patterns are similar from day to day), the scientists determined that male flies have more rhythmicity within their circadian patterns compared to females.
Sex differences were also detected when social interaction was a factor. Previous studies had shown that a fly had increased daytime sleep if it was exposed to 30 same-sex flies for five days compared to being kept in isolation. Harbison discovered that males were more sensitive to this paradigm than females; almost all the males in social groups had increased daytime sleep whereas the female response was more varied.
Harbison’s studies established that there are indeed differences in sleep between male and female fruit flies. Genome-wide association studies (GWAS) have investigated 1.9 million variants for association with sleep in flies; three studies have found thousands of genes. But if only one sex was focused on, 17-35 percent of genes would have been missed! She did note that she would like to investigate gene expression in the fly brain to determine if the same differences occur there as at the whole-body level.
Fruit flies are invertebrates and therefore not covered by the NIH policy that requires sex as a biological variable to be factored into research using vertebrate animals and in human studies. But Harbison urged that unless there is a compelling reason to not look at both sexes, it should be done…even in fruit flies!
The aim of SGHD SIG is to promote information exchange and interaction among NIH scientists who work on or are interested in aspects of sex-based research in any segment of the research continuum or in sex differences research relevant to health and disease as approached via a wide variety of scientific disciplines; to serve as an open forum for the presentation and discussion of the application of sex as a biological variable and aspects of sex-related research, including sex differences in mechanisms underlying health and disease; and to foster and support networking between NIH and non-NIH researchers focusing on the investigation of sex and gender influences. For more information, visit https://oir.nih.gov/sigs/sex-gender-health-disease or contact the co-chairs: Elena Gorodetsky (egorod@mail.nih.gov) or Katrina Serrano (katrina.serrano@nih.gov).
MPIG and SBIG: Determining Molecular Structures
The Membrane Protein Interest Group (MPIG) and the Structural Biology Interest Group (SBIG) are science hubs for brainstorming interactions among scientists who strive to determine molecular structures by applying the principles of computational biology and biophysics and using experimental tools such as X-ray crystallography, electron microscopy (EM), mass spectrometry, nuclear magnetic resonance (NMR), cryo-electron microscopy (cryo-EM), and computational approaches.
The SBIG/MPIG Student and Postdoc Symposium, held at the NIH Cloisters (Building 60) on May 24, showcased the novel discoveries and significant contributions made by the research groups from several institutes.
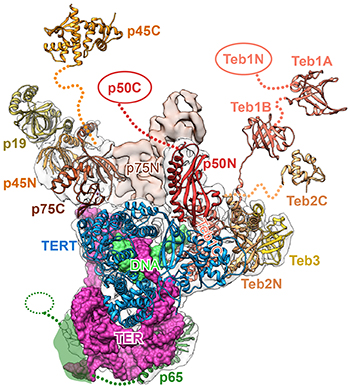
CREDIT: JIANSEN JIANG, NHLBI
In his keynote talk, Jiansen Jiang (NHLBI) described how cryo-EM revealed structures of telomerase and provided new insights into telomeric DNA synthesis. Shown: A map of the ribbon structure of a telomerase complex.
Many of the researchers in the SBIG/MPIG use cryo-EM, an EM technique in which a sample is cooled to extremely low temperatures. Cryo-EM helps scientists decipher and visualize the structural properties of a variety of protein molecules including the human anion exchanger 1, the major membrane protein in red blood cells that plays a key role in carbon dioxide exchange in the blood; tubulin isoform composition and microtubule dynamics; Ton, a multi-subunit membrane protein complex and molecular engine found in the inner membrane of gram-negative bacteria; voltage-activated potassium channels in lipid nanodiscs (synthetic model membrane systems); particles of virus-associated RNA-I/protein kinase RNA-activated PKR inhibiting; transposons widely used for genomic engineering; the outer mitochondrial membrane sorting and assembly machinery complex; and a membrane enzyme that serves as a basis for designing in silico inhibitors.
The symposium’s keynote speaker, Jiansen Jiang, who joined the National Heart, Lung, and Blood Institute (NHLBI) as an investigator in October 2017, spoke about how cryo-EM structures of telomerase, a critical determinant of human health—affecting aging, cancer, and stem-cell renewal—can reveal new insights into telomeric DNA synthesis.
Structural biology has also paved way for phenomenal contributions in the discovery of novel therapeutics. One such noteworthy discovery is how proton pump inhibitors—traditionally used to treat gastrointestinal tract bleeding, stomach ulcers, and acid reflux—could be also used as inhibitors of HIV-1. NHLBI postdoc Madeleine Strickland explained how her group used NMR to determine the high-resolution structure of one of the inhibitors, tenatoprazole, in complex with Tsg101 (a protein that seems to play a role in the pathogenesis of HIV). Understanding the structure of tenatoprazole will form the basis for the further development of a new class of HIV-1 inhibitor.
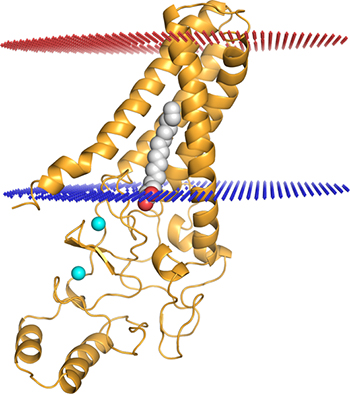
CREDIT: MITRA RANA, NICHD
Mitra Rana (NICHD) highlighted the importance of understanding the structure of DHHC enzymes, which are involved in many cellular processes, including cancer,and modify other proteins. Shown: DHHC20 palmitoyltransferase enzyme (gold) embedded into a modeled lipid bilayer (red and blue lines); two zinc ions (cyan spheres) bound by the enzyme; and a palmitate lipid chain (gray) covalently attached to the enzyme’s active site.
Other work highlighted included that of visiting fellow, Mitra Rana (National Institute of Child Health and Human Development), who gave a talk on the structural basis of fatty acyl recognition and transfer by the DHHC20 palmitoyltransferase enzyme. DHHC enzymes, involved in many cellular processes including cancer, modify other proteins by attaching fatty acids, usually palmitate, to them. In addition, Herve Celia (National Institute of Diabetes and Digestive and Kidney Diseases) discussed the structural investigation of Ton, a membrane molecular motor. In gram-negative bacteria—such as Escherichia coli, Salmonella, and Shigella—outer membrane transporters import nutrients by coupling to an inner membrane protein complex called the Ton complex.
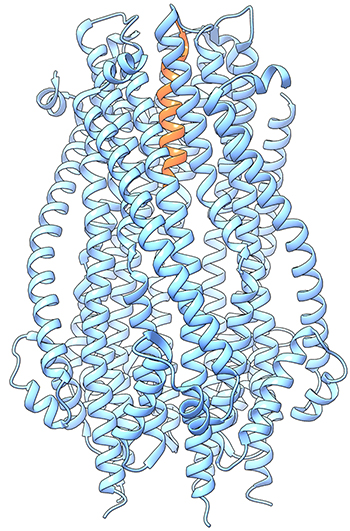
CREDIT: HERVE CELIA, NIDDK
Herve Celia (NIDDK) discussed the structural investigation of Ton, a membrane molecular motor. Shown: crystallographic structure of two subunits of the Ton complex.
Scientists also rely on computer simulations to decipher the complexities of molecular structures. Research Fellow Vanessa Leone (National Institute of Neurological Disorders and Stroke) described how her group used computer simulations to facilitate an integrative, molecular-level interpretation of structural data (through X-ray crystallography) and spectroscopic data (through double electron-electron resonance spectroscopy) on membrane proteins. In particular, her group looked at a glycine betaine symporter (BetP), which regulates the cell’s response to osmotic stress (efflux or influx of water) of Corynebacterium glutamicum, a soil bacterium used extensively in biotechnology. Her analysis showed that BetP’s molecular structure goes through a series of changes during its transport cycle. For example, BetP adopts an inward-facing conformation in the presence of excess sodium and other conformations in the presence of other substrates.
In addition, there were more than 50 posters at the symposium. One of the posters, presented by postdoc Amila Abeykoon (National Institute of Diabetes and Digestive and Kidney Diseases) highlighted a collaboration with Susan Gottesman’s lab (National Cancer Institute) to find novel antibiotics to treat the antibiotic-resistant Klebsiella pneumoniae. The bacterium secretes a polysaccharide capsule, which protects it from the immune system and any antibiotics. The researchers are using high-resolution structural analysis of the proteins that regulate the capsule synthesis in hopes of gaining new insights that may lead to the development of novel antibiotics that can be used to treat K. pneumoniae infections.
Throughout the day, members of the SBIG/PMIG group demonstrated how structural biology provides a high-resolution view deep into the molecular architecture of cells, revealing the biological mechanisms that are fundamental to life. The researchers’ findings are key to many innovations in chemistry, biotechnology, and medicine such as engineered enzymes, new drugs, innovative vaccines, and novel biomaterials.
For more information on the Membrane Protein Interest Group (MPIG) contact Reinhard Grisshammer (240-781-4031 or reinhard.grisshammer2@nih.gov). For more information on the Structural Biology Interest Group (SBIG), go to https://oir.nih.gov/sigs/structural-biology-interest-group or contact Antonina Roll-Mecak (301-402-7353 or Antonina.roll-mecak@nih.gov) or Anirban Banerjee (301-496-4855 or anirban.banerjee@nih.gov).
This page was last updated on Thursday, April 7, 2022
