Research Briefs
Read about scientific advances and discoveries by NIH intramural scientists: New app streamlines genome-sequencing data; biomarker predicts neurodegenerative disease; language shapes how humans interpret colors; antibodies neutralize immunodeficiency virus; dual-role cytokine mediates allergy; and mechanisms revealed behind DNA repair.
NCI: SHERLOCK-GENOME MAKES WHOLE GENOME SEQUENCING DATA USER-FRIENDLY
BMC GENOMICS, NCI
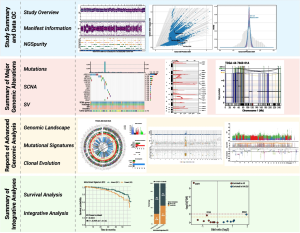
The illustration shows an overview of the four major modules and example visualizations supported by Sherlock-Genome to manage and present whole genome sequencing data in a user-friendly manner.
Comprehensive analysis of an individual’s entire genome via whole genome sequencing (WGS) is far from elementary. Sherlock-Genome, a new interactive web application developed by NCI scientists, aims to streamline how such complex data are analyzed, integrated, shared with others, and visually presented. Among its many uses, WGS has significantly contributed to advancements in the field of cancer genomics. For example, it can precisely detect genetic mutations and complex genomic alterations within a tumor, which can then be used for a personalized approach to cancer treatment.
However, the complexity of WGS data poses significant challenges, especially for researchers who don’t have a background in bioinformatics or strong programming skills. The NCI team developed Sherlock-Genome to simplify the analysis and visualization of WGS data and to make cancer genomic research more accessible to a wider range of scientists. “It also facilitates the integration of genomic findings with clinical data, bridging the gap between research and patient care,” said Tongwu Zhang, a Stadtman investigator who led the study.
Sherlock-Genome users can upload and analyze WGS results, integrate their data with other datasets, such as from clinical and epidemiological studies, and generate publication-ready figures. The platform incorporates four major modules for cancer genomic analysis including study summary data, summary of major genomic alterations, reports of advanced genomic analysis, and summary of integrative analyses.
“Sherlock-Genome marks a significant step toward making complex genomic analyses more accessible and interpretable, ultimately advancing human health research,” said Zhang. Researchers may access the source code and installation instructions for Sherlock-Genome on Github. (NIH authors: A. Klein, J. Zhong, M.T. Landi, and T. Zhang, PMID: 39819269)
[BY JOHN CARLO J. COMBISTA, NIMH]
NINDS, NIAID: CEREBROSPINAL FLUID BIOMARKER PREDICTS LEWY BODY DISEASES
Early detection of a protein misfolding and aggregation process can act as a screening tool by identifying individuals who might later develop a central Lewy body disease (LBD). The findings come from a prospective longitudinal study at the NIH CC conducted by investigators at NINDS and NIAID’s Rocky Mountain Laboratories. LBDs affect more than 1 million people in the United States and are associated with the buildup of abnormal alpha-synuclein proteins in the brain that results in progressive movement disorders and dementia.
One of the factors associated with LBDs, alpha-synuclein seeding activity (SSA), is a process by which a sample of a patient’s biofluid triggers alpha-synuclein in a test tube to polymerize and misfold. It is widely thought that the triggering factor in the biofluid is itself misfolded alpha-synuclein. Small chains (oligomers) of alpha-synuclein protein molecules or misfolded alpha-synuclein damages neurons that use catecholamines such as dopamine as the chemical messenger, resulting in manifestations such as Parkinsonism or cognitive dysfunction.
Using an alpha-synuclein seed-amplification assay, the researchers analyzed cerebrospinal fluid from individuals with self-reported risk factors for LBD and followed them for up to 7.5 years. They found that 64% of participants with increased SSA went on to develop an LBD compared with 5% who fell below a cutoff value for SSA activity. Other findings from the study indicated that LBDs involve multiple functional abnormalities in the central nervous system, suggesting targets for future research aimed at delaying or even preventing symptomatic disease.
The addition of multiple tests such as cardiac biomarker assessment by positron-emission scans may prove even more useful. “In at-risk individuals, a combined biomarkers approach that includes cerebrospinal SSA may predict accurately who will go on to develop a symptomatic central LBD during the subsequent several years,” said David Goldstein, first author on the study. (NIH authors: D.S. Goldstein, P. Alam, P. Sullivan, C. Holmes, J. Gelsomino, A.G. Hughson, and B. Caughey, PMID: 39817492)
NIAID: BROADLY NEUTRALIZING ANTIBODIES AGAINST SIV IDENTIFIED

CELL REPORTS, NIAID
Four newly discovered lineages of antibodies broadly neutralized simian immunodeficiency virus by binding to the MPER, a region of the virus that could be targeted in vaccine development against the closely related human immunodeficiency virus.
Researchers at NIAID’s Vaccine Research Center discovered and characterized four lineages of broadly neutralizing antibodies against simian immunodeficiency virus (SIV), expanding scientists’ toolkit to design a critically needed vaccine against the closely related HIV. Nearly 40 million people worldwide currently live with HIV, the virus responsible for AIDS.
The scientists homed in on an area of SIV known as the membrane proximal external region (MPER), which allows the virus to merge with and infect a host cell. MPER has been shown in human studies to be a target for broadly neutralizing antibodies against HIV. By screening plasma of multiple SIV-infected rhesus macaques, the investigators identified two animals that could effectively neutralize SIV and isolated 13 antibodies from those animals with specificity to the MPER. The antibodies were found to belong to four distinct, genetically similar lineages that bound in a similar way as broadly neutralizing antibodies against the HIV MPER protein found in humans.
According to Jason Gorman, first author on the study, “The results here expand the availability of broad SIV antibody reagents to help facilitate use of the SIV nonhuman primate animal model for studying HIV and can help inform HIV antigen design for vaccines targeting the MPER.” (NIH authors: J. Gorman, D. Renguang, Y. Lai, A.S. Mohammed, H.A.D. King, K. Song, K. Manalang, C.A. Gonelli, C.A. Schramm, C. Cheng, R. Nguyen, D. Ambrozak, A. Druz, C. Shen, Y. Yang, D.C. Douek, P.D. Kwong, M. Roederer, and R.D. Mason, PMID: 39792559)
[BY TAYLOR FARLEY, NIAID]
NHLBI, NIAID: IMMUNE CYTOKINE SERVES DUAL ROLE IN MEDIATING ALLERGIC RESPONSE
Scientists from NHLBI and NIAID have identified a dual role for thymic stromal lymphopoietin (TSLP), an immune cytokine that can both promote and limit inflammation in type 2 immunity, providing new insight into how the immune response is balanced. The mechanisms behind type 2 immunity are not fully understood and involve a set of inflammation-driven pathways that when dysregulated can contribute to allergic reactions such as asthma.
TSLP is known to activate type 2 helper T (Th2) cells, the immune system’s frontline responders that produce type 2 inflammatory cytokines and can also trigger symptoms such as swelling and airway constriction. Blocking that mechanism is behind how current anti-TSLP therapies used to treat severe asthma are thought to work.
In their study, the investigators found that besides its action on Th2 cells, TSLP also stabilizes regulatory T cells (Tregs), which act as the immune system’s peacekeepers to prevent excessive inflammation. Using a mouse model, the researchers genetically deleted the TSLP receptors from both types of T cells, which, as expected, resulted in a muted immune response to an airway inflammation challenge. However, when TSLP receptors were selectively removed from Tregs only, the immune system became dysregulated. Lacking TSLP’s stabilizing influence, Tregs not only failed to suppress inflammation but also began driving the process and exacerbated allergic responses. Conversely, when TSLP signaling in Tregs remained intact, the cells maintained their immunosuppressive identity and ability to limit allergic inflammation.
According to the authors, “The results expand the known roles for TSLP and indicate additional points at which TSLP might be targeted to modulate the immune response.” (NIH authors: R.K. Gurram (now at AstraZeneca), P. Li, J. Oh, X. Chen, R. Spolski, X. Yao, J.X. Lin, S. Roy, M.J. Liao, C. Liu, Z.X. Yu, S.J. Levine, J. Zhu, and W.J. Leonard, PMID: 39792638)
[BY ARWAA MEHRAN, NICHD]
NCI, NCATS: NEWLY IDENTIFIED PROTEINS AND PATHWAYS MAINTAIN GENOMIC STABILITY
SCIENCE ADVANCES, NCI
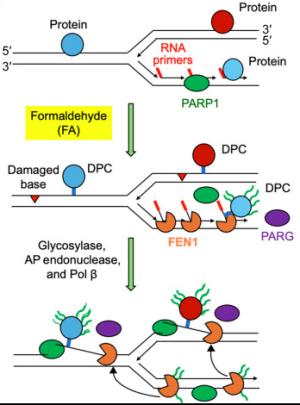
Illustration showing how a process known as PARylation uses the PARP1 enzyme to mark damaged DNA sites and recruits the FEN1 protein to initiate repair.
DNA-altering environmental pollutants and internal metabolic byproducts, such as formaldehyde, can lead to the development of diseases such as immunodeficiencies, neurodegeneration, and cancer. Research led by scientists at NCI, NCATS, and the University of Maryland (Baltimore) identified the proteins involved in formaldehyde-induced DNA-protein cross-links (DPCs), a common type of DNA lesion, and discovered a new molecular pathway by which those lesions are repaired.
In a series of experiments, the researchers revealed that the protein FEN1 excises the helix-distorting flap structures that are a hallmark of DPC damage. FEN1 was also found to repair another type of DPC caused by inhibition of the TOP2 enzyme, commonly associated with chemotherapy drugs. They found that FEN1 works through a DNA repair mechanism regulated by a process known as PARylation, which uses the PARP1 enzyme to tag damaged sites in the genome and recruits FEN1 to initiate repair. In their paper, the authors note that the findings “provide previously unrecognized pieces to the puzzle of DPC repair and a molecular foundation for the etiology of DPC-induced diseases.” (NIH authors: Y. Sun, L.M. Jenkins, L.H. El Touny, X. Yang, U. Jo, T.K. Maity, L.K. Saha, S. Saha, K. Cheng, and Y. Pommier, PMID: 39792662)
[BY CASEY CARGILL, NEI]
This page was last updated on Wednesday, March 19, 2025
