Research Briefs
Read about NIH scientific advances and discoveries by intramural scientists:
- NIEHS: FOXL2 LINKED TO ENDOMETRIOSIS, FEMALE INFERTILITY, AND CANCER
- NHLBI, NCI: NEW BLOOD TEST FOR THE EARLY DETECTION OF ACUTE HEART-TRANSPLANT REJECTION
- NIDA: NATIONWIDE INCREASE IN METHAMPHETAMINE OVERDOSE DEATHS
- NIAID (RML): SALMONELLA SWIMMING BEHAVIOR MAY PROVIDE CLUES TO INFECTION
- NIDDK, CC, NINR, NIAAA: NIH RESEARCHERS COMPARE LOW-FAT AND LOW-CARB DIETS
- NIAID, NIGMS, NCI, NIDDK, NHGRI: HOW THE GUT MICROBIOTA IS TRAINED TO PREVENT INFECTIONS
NIEHS: FOXL2 LINKED TO ENDOMETRIOSIS, FEMALE INFERTILITY, AND CANCER
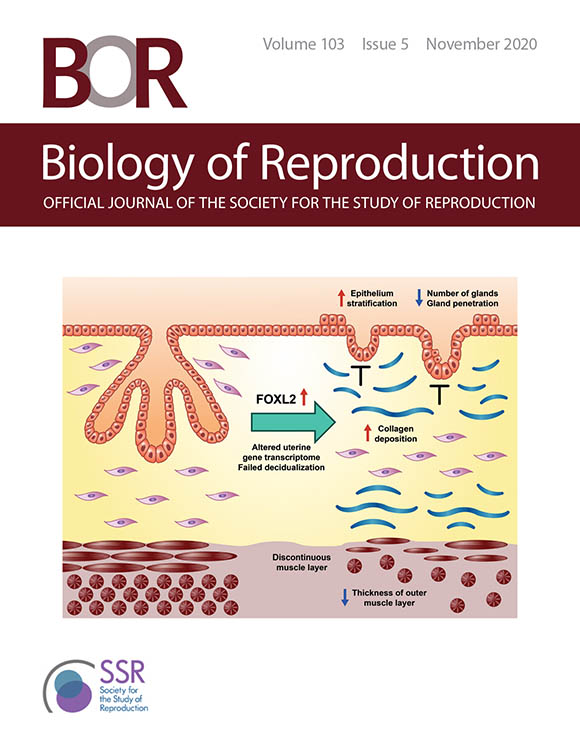
CREDIT: IMAGE USED WITH PERMISSION FROM THE SOCIETY FOR THE STUDY OF REPRODUCTION
The cover of the November 2020 issue of Biology of Reproduction features a graphical abstract of the first study. Shown: FOXL2 overexpression in the uterus induced epithelial stratification, blunted adenogenesis (development of uterine glands), increased fibrosis, and disrupted myometrium (muscular outer layer of the uterus) leading to impaired decidual responses (changes in endometrial cells in preparation for pregnancy).
Two teams of NIEHS authors published companion articles that further underscored the importance of the transcription factor forkhead box L2 (FOXL) in women’s ovarian physiology and fertility and clarified the role of excess FOXL2 in endometriosis (a disorder in which endometrial tissue, which normally lines the uterus, grows abnormally outside of the uterus). FOXL2 regulates sex differentiation and reproductive function.
In humans, FOXL2 genes that mutate or lose function are associated with premature infertility and ovarian cancer. Expression of FOXL2 is increased in the uterine tissue of women with endometriosis, which can be influenced by endocrine disruptors in the environment. In the first study, the research team generated mice with overexpression of FOXL2 and reported that too much FOXL2 altered differentiation and function of uterine epithelial cells. In addition, endometrial stromal cells showed increased fibrosis, interfering with development of the endometrial glands. The study identified mouse uterine pathways regulated by FOXL2 that are similar to pathways elevated in uterine samples of women with endometriosis, suggesting the potential for better diagnostic and therapeutic tools to treat endometriosis, a widespread disease.
In the second study, also based on a mouse model, the researchers evaluated the effects of irregular FOXL2 activity on female reproductive physiology and disorders. By studying its effects on ovarian development and function from the fetal stage through adulthood, the scientists discovered that excessive FOXL2 production made it harder for certain cells to differentiate, and the follicles did not form correctly. The female mice could not ovulate and were infertile. This study reveals that fine-tuned production of FOXL2 is absolutely essential for proper ovarian physiology and fertility. This potential cause of ovarian disorders in women may point to treatment targets. (Study 1: NIH authors: R. Li, S.-P. Wu, L. Zhou, B. Nicol, H.H.-C. Yao, and F.J. DeMayo, Biol Reprod 103:951–965, 2020; DOI:10.1093/biolre/ioaa143. Study 2: NIH authors: B. Nicol, K. Rodriguez, and H.H.-C. Yao, Biol Reprod 103:966–977, 2020; DOI:10.1093/biolre/ioaa146.
NHLBI, NCI: NEW BLOOD TEST FOR THE EARLY DETECTION OF ACUTE HEART-TRANSPLANT REJECTION
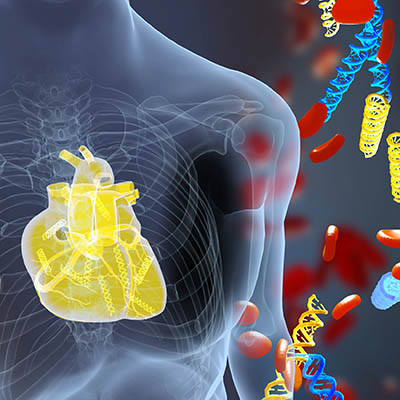
CREDIT: ERINA HE, NIH MEDICAL ARTS
NHLBI: A new blood test measures donor DNA fragments and detects acute heart-transplant rejection earlier than current methods. The illustration shows DNA fragments (yellow) derived from a transplanted heart alongside the patient’s own DNA (blue).
Researchers at NHLBI have developed a blood test for the early detection of acute heart-transplant rejection, a potentially fatal condition that can occur within a few months after receiving a heart transplant. In an article published in Circulation, the authors including Sean Agbor-Enoh, chief of NHLBI’s Laboratory of Applied Precision Omics, estimated that by using this new test, doctors can safely avoid 81% of invasive heart-tissue biopsies, the current method used to detect rejection. In this procedure, a catheter is inserted into an incision in the neck, then threaded through a blood vessel and into the heart chamber to obtain a tissue sample, which is later analyzed for signs of acute rejection.
The new blood test involves a quick blood draw and measures donor-derived cell-free DNA (ddcfDNA), short fragments of DNA released by donor heart tissue into the bloodstream if a recipient’s immune system attacks the heart. In their study, the researchers analyzed the blood concentrations of ddcfDNA in 165 heart-transplant recipients (out of 171 participants), finding higher concentrations of ddcfDNA in patients with acute rejection. Importantly, the researchers found that ddcfDNA is highly sensitive in detecting acute rejection and that its performance was better than that of heart biopsies. The new blood test may be able to detect rejection as early as 28 days after a heart transplant; it can take weeks or months to detect rejection using the current methods. (NIH authors: S. Agbor-Enoh, C. Mutebi, K. Yu, P. Shag, I. Tunc, S. Hsu, H. Kong, A. Bikineyeva, A. Marishtam, K. Bhatti, Y. Yang, M.K. Jang, and H.A. Valantine, Circulation 2021; DOI:10.1161/CIRCULATIONAHA.120.049098)
NIDA: NATIONWIDE INCREASE IN METHAMPHETAMINE OVERDOSE DEATHS
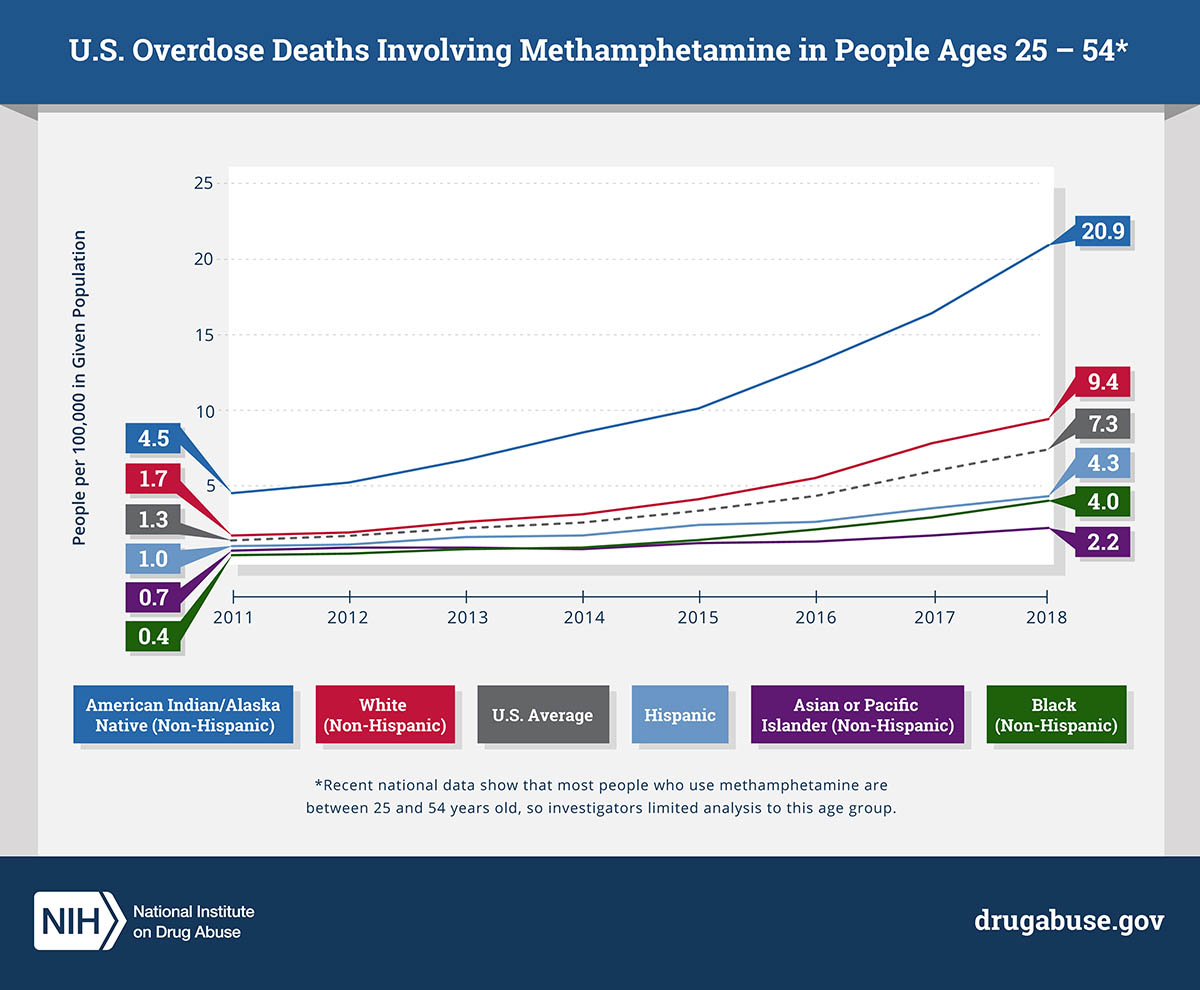
CREDIT: NIDA
NIDA: Methamphetamine overdose deaths in people aged 25 to 54 increased from 2011 to 2018, especially among non-Hispanic American Indians and Alaska Natives.
In a study published in JAMA Psychiatry, researchers at NIDA found a more than fivefold increase in methamphetamine-involved age-adjusted overdose death rates over an eight-year period from 2011 to 2018, especially among American Indians and Alaska Natives. The analysis, based on data from the CDC’s National Vital Statistics System, examined overdose death rates of individuals aged 25–54 by gender and race/ethnicity. The researchers found that overdose death rates rose across all racial and ethnic groups and among both men and women, although men had significantly higher rates than women within each racial/ethnic group. American Indians and Alaska Natives had by far the highest death rates, which more than quadrupled over the study period. “Identifying populations that have a higher rate of methamphetamine overdose is a crucial step toward curbing the underlying methamphetamine crisis,” said Beth Han, lead author on the study. “By focusing on the unique and culturally specific needs of individuals, we can begin to move away from one-size-fits-all approaches and toward more effective, tailored interventions.” (NIH authors: B. Han, J. Cotto, K. Etz, E.B. Einstein, W.M. Compton, and N.D. Volkow, JAMA Psychiatry 2021; DOI:10.1001/jamapsychiatry.2020.4321)
NIAID (RML): SALMONELLA SWIMMING BEHAVIOR MAY PROVIDE CLUES TO INFECTION
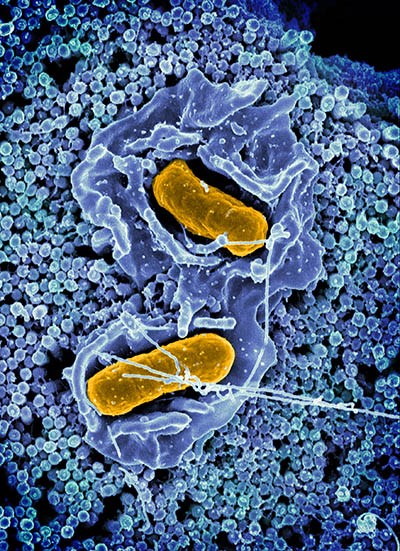
CREDIT: ELIZABETH R. FISCHER AND AUSTIN ATHMAN (COLORIZED), RML, NIAID
A scanning electron micrograph of Salmonella enterica serovar Typhimurium (yellow), a common cause of foodborne infection, invading a human epithelial cell (blue).
Scientists know a lot about Salmonella enterica serovar Typhimurium (S. Typhimurium), one of the most common causes of foodborne infection, and how these flagella-propelled bacteria swim in short, random spurts. Yet, until now, they didn’t understand how the type of swimming can contribute to infection of the epithelial cells lining the inside of the intestine. Researchers at NIAID’s Rocky Mountain Laboratories (Hamilton, Montana) recently discovered that a certain protein can turn S. Typhimurium bacteria in the gut into tiny missiles that swim straight.
In this study, the authors identified the protein McpC, short for methyl-accepting chemotaxis protein C, that causes the bacteria to swim straight when they are ready to infect cells and promotes invasion of surface epithelial cells in the gut. “In the presence of McpC, the bacteria swim in longer, straighter runs than McpC–defective mutants,” said Olivia Steele-Mortimer, senior author on the study. “This [longer, straighter run] is tightly coupled to invasion in the gut.”
McpC could be a potential target for developing new antibacterial treatments to hinder the ability of S. Typhimurium to infect and colonize the gut, according to the study authors. The researchers hypothesize that controlled smooth swimming could be a widespread bacterial infection strategy and that their findings could result in the development of novel antibiotics. (NIH authors: K.G. Cooper, A. Chong, L. Kari, B. Jeffrey, T. Starr, C. Martens, and O. Steele-Mortimer, Nat Commun 12:article number 348, 2021; DOI:10.1038/s41467-020-20558-6)
NIDDK, CC, NINR, NIAAA: NIH RESEARCHERS COMPARE LOW-FAT AND LOW-CARB DIETS
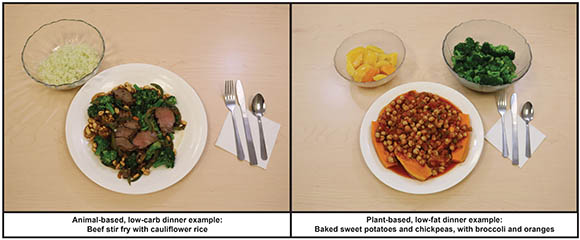
CREDIT: AMBER COURVILLE AND PAULE JOSEPH, NIH
Example of foods provided to study participants: animal-based diet (left) and plant-based diet (right).
NIDDK scientists led a small, highly controlled, inpatient study to compare the effects of a low-fat, high-carbohydrate, plant-based diet with a high-fat, low-carbohydrate, animal-based diet. The study sought to compare calorie (or energy) intake, hormone concentrations, body weight, and satiety levels. A total of 20 adults (11 men and nine women), housed in the NIH Clinical Center’s Metabolic Research Unit, were given one diet for two weeks, immediately followed by two weeks on the other diet. All participants were given three meals a day plus snacks and allowed to eat as much as they desired.
High-carbohydrate foods were expected to increase hunger and result in excess calorie intake because they can cause large swings in blood glucose and insulin.
But the results were surprising: When people were on the plant-based diet, they ate 550–700 fewer calories daily, but had higher insulin and blood glucose concentrations than when they ate the animal-based diet. Despite differences in calorie intake, there were no significant differences in terms of participant rating of hunger, enjoyment of meals, or feeling full. Both diets also resulted in weight loss, although this loss was only significant for the plant-based diet during the first week.
The findings suggest that the factors resulting in overeating and weight gain are more complex than the amount of carbs or fat in one’s diet. In fact, there could be benefits to both diets if only in the short term.
“While the low-fat, plant-based diet helps curb appetite, the animal-based, low-carb diet resulted in lower and more steady insulin and glucose levels,” said NIDDK Senior Investigator Kevin Hall, the study’s lead author, in a news release. “We don’t yet know if these differences would be sustained over the long term.” (NIH authors: K.D. Hall, J. Guo, A.B. Courville, J. Boring, R. Brychta, K.Y. Chen, V. Darcey, A.M. Gharib, I. Gallagher, R. Howard, P.V. Joseph, L. Milley, R. Ouwerkerk, K. Raisinger, I. Rozga, A. Schick, M. Stagliano, S. Torres, M. Walter, P. Walter, S. Yang, and S.T. Chung, Nat Med 2021; DOI:10.1038/s41591-020-01209-1)
NIAID, NIGMS, NCI, NIDDK, NHGRI: HOW THE GUT MICROBIOTA IS TRAINED TO PREVENT INFECTIONS
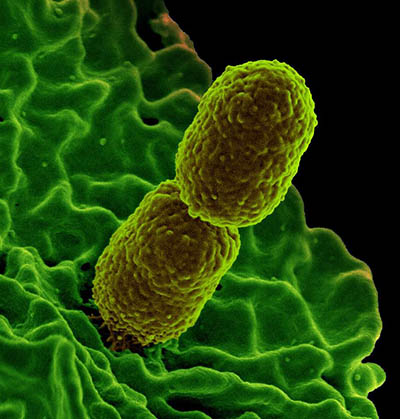
CREDIT; KEN PEKOC, NIAID
Colorized scanning electron micrograph showing carbapenem-resistant Klebsiella pneumoniae interacting with a human neutrophil.
In an NIAID-led study, NIH scientists have made a quantum leap in our understanding of ways the gut microbiota, the community of diverse microbes naturally living in the gastrointestinal tract, wards off bacterial infections. They identified a nutrient—taurine—that helps the gut recall prior infections and kill invading bacteria, such as Klebsiella pneumoniae (Kpn).
The gut microbiota can shield us from Kpn and other bacterial infections via a process known as colonization resistance. To uncover the mysterious mechanism of colonization resistance, the NIH researchers used two groups of pathogen-free mice—one harboring the gut microbiota of mice from the wild and the other experiencing a transient infection with an attenuated strain of the food-borne pathogen Yersinia pseudotuberculosis. They found that both groups of mice showed enhanced resistance to Kpn infections, demonstrating that the microbiota that has already experienced an infection helps resist a subsequent infection.
The researchers discovered that the prior infection resulted in the proliferation of a class of taurine-utilizing bacteria called Deltaproteobacteria in the gut. Taurine, an amino acid, helps the body digest fats and oils and is naturally found in bile acids in the gut. Remarkably, taurine given to mice as a supplement in drinking water was sufficient to train the microbiota to fight off Kpn infections. Further experiments revealed that poisonous hydrogen sulfide gas released as a byproduct of taurine promoted enhanced resistance, possibly by inhibiting pathogen respiration. This study may aid in identifying microbiota-based alternatives to antibiotics for fighting off life-threatening bacterial infections. (NIH authors: A. Stacy, V. Andrade-Oliveira, J.A. McCulloch, B. Hild, J.H. Oh, P.J. Perez-Chaparro, C.K. Sim, A.I. Lim, V.M. Link, M. Enamorado, G. Trinchieri, J.A. Segre, B. Rehermann, and Y. Belkaid, Cell 184:615–627.e17, 2021; DOI:10.1016/j.cell.2020.12.011)
This page was last updated on Tuesday, February 15, 2022
