News You Can Use
NIH’s Top-Ranking Supercomputer
The Computational Behemoth, Biowulf

PHOTO BY BEN CHAMBERS, OIR
Combined, Biowulf’s 10 high-performance storage systems, two of which are pictured here (each occupies two racks), provide more than 25 petabytes of storage capacity.
Tucked away behind the nondescript walls of Building 12 lies a computational behemoth known as Biowulf. The state-of-the-art supercomputer enables scientists in the NIH Intramural Research Program (IRP) to analyze massive datasets and attempt projects whose sheer scale would make them otherwise impossible.
Although the intramural community has harnessed the power of supercomputers since 1986, Biowulf itself was first launched in 1999. The size and complexity of scientific datasets were growing rapidly then, in no small part due to the massive amounts of data being produced by the Human Genome Project. More recently, modern scientific endeavors in fields such as biochemistry, microbiology, molecular dynamics, genomics, and biomedical imaging have once again dramatically increased scientists’ computational needs.
To meet the demand, in 2014 the NIH embarked on a five-year initiative to dramatically enhance Biowulf’s already considerable computing power. Four years in, the supercomputer has become a world-class resource used by more than one-third of the thousand-plus IRP research labs.

PHOTO BY BEN CHAMBERS, OIR
Fiber optic cables support high-capacity, high-speed data transfer.
In 2017, Biowulf became the first machine completely dedicated to advancing biomedical research to break into the top 100 of TOP500.org’s list of the most powerful supercomputers in the world, landing at number 66 in the rankings. Since 2014, the number of active Biowulf users has doubled (from 640 to 1,276), and jobs submitted have increased more than 800-fold (from 3.7 million to 32.1 million). More than 2,000 journal articles have been published based on data analyzed using Biowulf.
With Biowulf at their service, intramural researchers can process large-scale datasets much faster than would be possible with standard computational equipment. The revamped supercomputer has been a boon to researchers all across the NIH, contributing to publications on topics ranging from how the time of day affects the brain’s water content, to how genes influence sleep duration in fruit flies, to a hunt for antibiotic-resistant bacteria in hospital plumbing.

PHOTO BY BEN CHAMBERS, OIR
Biowulf requires massive cooling support and its under-floor chilled water lines are easily accessible for maintenance.
The system is also being used to aid machine-learning projects, as researchers attempt to develop algorithms that can rapidly and accurately trace the borders of cells in a microscope image or diagnose illnesses by looking at magnetic-resonance-imaging scans or other scans. Overall, about a third of Biowulf’s computing power is used for computational chemistry, a third for structural biology, and 20 percent for genomics, with dozens of other areas of biomedical inquiry using the remainder.
Biowulf’s ascendance represents remarkable progress for both the NIH and scientific research generally, said Andy Baxevanis, Director of Computational Biology for the NIH Office of Intramural Research. “High-performance computing is a critical element of modern-day biomedical research and a vital resource for the intramural community,” he said. “Biowulf’s expansion is a sign that the NIH recognizes the importance of investing in state-of-the-art technologies to permit our scientists to remain at the forefront of their fields.”
The High-Performance Computing (HPC) Group at NIH’s Center for Information Technology (CIT) manages Biowulf and supports its users. The name “Biowulf” is a play on words combining “biology” with the name of a specific type of HPC system called a Beowulf cluster (Beowulf was named after the hero in the Old English epic poem, who had “thirty men’s heft of grasp in the gripe of his hand.”) NIH’s HPC Group first learned about the Beowulf-cluster approach to supercomputing from its creator, then–NASA Goddard Space Flight Center Senior Scientist and current Indiana University Professor Tom Sterling, at a 1997 conference in San Jose, California. Each cluster is composed of several low-cost computers, called nodes, connected to one another and to other clusters via a centralized network and loaded with open-source software. By connecting enough nodes together, Beowulf systems can generate immense computing power for a relatively low cost.
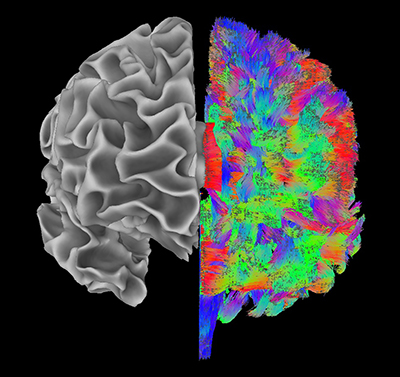
IMAGE BY SCIENTIFIC AND STATISTICAL COMPUTING CORE (“AFNI SOFTWARE GROUP”), NIMH
Different aspects of brain structure are visible in this image, produced using Biowulf. The left hemisphere shows the boundary of gray and white matter tissue, and the linear structures in the right hemisphere are computational estimates of where white matter might be connecting different parts of the brain.
Today, Biowulf is composed of more than 4,200 nodes collectively containing 97,000 cores, each of which can independently run program code. The computer system provides over 25 petabytes of primary storage to accommodate the large numbers of simultaneous jobs and large-scale distributed memory tasks common in the biosciences. To put that into perspective, Biowulf’s primary storage could hold the information found on roughly 12.5 trillion pages of standard printed text—the equivalent of more than 24 billion copies of Charles Darwin’s On the Origin of Species. Additionally, Biowulf has 528 graphical processing units that accelerate data analysis by handling multiple computational tasks simultaneously. The system also supports over 600 scientific programs, packages, and databases, many of which are custom-built to support the biosciences.
The HPC team was awarded the NIH Director’s Award in 2017 for its incredible work in building up the Biowulf system. Yet, even with its astounding computing power, the supercomputer can barely keep up with the ever-expanding demands of IRP researchers. The fifth phase of the expansion effort, which will come online in July 2019, will add another 30,000 cores to the system.
Andrea Norris, director of CIT and NIH’s chief information officer, looks forward to seeing more researchers realize the benefits of Biowulf. “We’re very excited about the research that intramural scientists have been able to perform using Biowulf,” she said. “NIH’s strategic investments in high-performance computing, its high bandwidth network, and other key technology capabilities are enabling the intramural community to achieve scientific discoveries that were previously beyond its reach.”
For more information on Biowulf, visit http://hpc.nih.gov or contact staff@hpc.nih.gov to see how Biowulf can help you advance your own research projects.
This page was last updated on Thursday, April 7, 2022
