Genome Editing
Eugene Koonin’s Continued Role in CRISPR, by Way of Yogurt
If Streptococcus thermophilus could erect statues, they’d do so to Eugene Koonin, a man responsible for saving untold trillions of their yogurt-making kind.
An evolutionary biologist and senior investigator in the National Library of Medicine’s National Center for Biotechnology Information (NCBI), Koonin hypothesized in 2005 that “spacer DNA” in the clustered regularly interspaced short palindromic repeats (CRISPR) loci of bacteria and archaea, which matched sequences of bacteriophages, could be a key part of a sort of adaptive immune system.
He didn’t know at the time that his hypothesis, which has since led to phage-resilient strains of S. thermophilus for the yogurt industry, would be a major step in creating a precision technique for editing genes in species as diverse as humans, whelk, and whales. But more on that later.
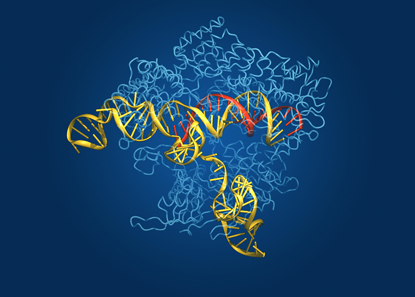
CREDIT: BANG WONG, BROAD INSTITUTE OF HARVARD AND MIT
Gene-Editing Technologies. Crystal structure of the CRISPR/Cas9 system, which enables DNA to be edited with unprecedented precision. Researchers use RNA guides called CRISPRs (red) to steer the Cas9 gene-editing enzyme (light blue) to a specific site on a DNA strand (yellow) that they wish to modify.
Koonin, then as now, was simply interested in “patterns, rules, and trends in the evolution of genomes,” he said. He and his colleagues were examining the genes adjacent to the CRISPR loci in bacteria and archaea as early as 2002 to identify partially conserved gene neighborhoods.
The CRISPR, with their spacers, were discovered in the late 1980s, but “no one knew what they were, and no one really cared,” Koonin said. Koonin figured the gene neighborhoods of CRISPR must have some important purpose from an evolutionary viewpoint, or they wouldn’t be so prominent. And so he probed.
Building on years of his own research on the concept of clusters of orthologous groups of proteins, Koonin and members of his NCBI lab, the Evolutionary Genomics Research Group, predicted that the functions of many of the proteins encoded by the genes in the CRISPR neighborhoods—known as CRISPR-associated proteins (Cas)—were to bind, cut, join, and unwind DNA and RNA.
That much would prove correct, as shown by extensive research over the following decade. To Koonin’s regret, however, his central hypothesis was off the mark. He had thought that the spacers were a novel system of DNA repair. He missed the fact that these genes were almost always adjacent to strange batteries of repeats . . . which would be so important to the discovery of the CRISPR method for editing genomes.
Nevertheless, the group published a paper about this in 2002 that garnered some attention.
It would take three years and three independent laboratories (two in France and one in Spain) to discover that some of the CRISPR spacers were homologous to bits of foreign DNA from bacteriophages or plasmid DNA. Those researchers suggested, in general terms, that the spaces might serve as a defense mechanism against infections.
Koonin and a senior member of his lab, Kira Makarova, read these studies in late 2005 and quickly connected all the dots. They developed a hypothetical scheme analogous to that for the system of RNA interference in eukaryotes—a premise both simple and radical—that prokaryote organisms could usurp fragments of enemy viruses by means of self-inoculation, to ensure that their progeny could recognize a future viral attack and dismantle it.
In short, it was adaptive immunity at a Lamarckian level (the heritability of acquired characteristics).

PHOTO CREDIT: YURI WOLF, NLM
Eugene Koonin
The paper Koonin’s group published in March 2006, with Makarova as the lead author, caught the attention of researchers at Danisco, an international company now owned by DuPont that provides enzymes and other biologics to food companies. The connection? S. thermophilus, the bacterium that can turn milk into yogurt, is something Danisco needs to thrive. Yet phages routinely infect and kill the bacteria.
Danisco wanted to test Koonin’s hypothesis. The scientists there exposed S. thermophilus to a phage; sure enough, while many perished, a few bacteria managed to inoculate themselves and lived and then passed that immunity on. The result has been a healthier strain of S. thermophilus, saving the company millions of dollars.
Danisco scientists, led by Rodolphe Barrangou and Philippe Horvath, published their results in the journal Science in March 2007. They had no idea that this natural inoculation, which has played out for eons, had any connection to gene editing. But other scientists soon would, and the result has been the development of the DNA-editing tool CRISPR-Cas.
How did bacteria and archaea pull off this feat of self-inoculation? That was the central question asked by the scientists who went on to establish the CRISPR-Cas gene-editing system, among them Emmanuelle Charpentier (currently at Max Planck Institute of Infection Biology in Berlin) and Jennifer Doudna (University of California, Berkeley), working together and separately, and also Feng Zhang (Massachusetts Institute of Technology, Cambridge, Massachusetts).
A Charpentier-led paper published in Nature in 2011, in particular, proposed that the enzyme Cas9 works with a stretch of RNA called trans-activating CRISPR RNA (tracrRNA) and other spacer-derived RNA called CRISPR RNA (crRNA) to snip the invading phage DNA and incorporate it into its own. By 2012, Charpentier and Doudna’s group published a paper in Science that described the method to use these RNA elements with Cas9 as the cleaving enzyme to find and cut DNA targets with unprecedented precision.
“The principle is very simple and extremely powerful,” Koonin said. It involves using “a piece of RNA to recognize in a highly specific manner a particular place in a DNA molecule and [cutting] the DNA in that place. If you think about that for a moment, this is the impossible dream of a genomic engineer.”
The CRISPR-Cas method was named one of the Science’s Top 10 Breakthroughs of 2013, just behind immunotherapy, another advance developed within the NIH intramural program.
Having instigated two milestones in the development of practical uses of CRISPR-Cas—prediction first of the functions of the Cas proteins and second of the mechanism of CRISPR-mediated adaptive immunity—and having saved some yogurt bacteria on the way, Koonin is back for what he, only half-jokingly, calls part three, noting that the Cas9 enzyme is just the tip of the iceberg.
In the past four years, he has published dozens of articles further defining CRISPR-Cas with lab colleagues Makarova, Yuri Wolf, Sergey Shmakov, and others. Key to their discoveries has been the creation of a pipeline to explore protein function computationally.
Koonin’s group identifies novel CRISPR-Cas loci and predicts protein function, and Zhang’s MIT lab tests these predictions experimentally, a hand-in-glove collaboration.
In October 2015, with MIT’s Zhang; Konstantin Severinov at Rutgers University in New Brunswick, New Jersey, and the Skolkovo Institute of Science and Technology in Moscow Oblast, Russia; and others, Koonin’s group published a paper in Molecular Cell detailing three more naturally occurring systems that show potential for genome editing. The proteins are provisionally termed class 2 candidates 1, 2, and 3 (C2c1, C2c2, and C2c3). Although they share properties with Cas9 and centromere and promotor factor–1 (Cpf1, another CRISPR nuclease that Koonin’s and Zhang’s groups discovered earlier in 2015), they have unique characteristics that may be exploited for novel genome-editing applications.
The situation “recapitulates what happened to restriction enzymes in the early days of genetic engineering in the 1970s,” Koonin said; then, the number of gene-editing tools grew dramatically. Koonin predicts that there are likely “a couple dozen” enzymes akin to Cas9 awaiting discovery, and his lab continues to hunt for them.
True to their research goals, Koonin and his colleagues have been able to infer an intricate evolutionary pathway for each these of defense systems.
Koonin doesn’t much care for yogurt, truth be told. He said vinification bacteria are more his speed, and indeed he was studying bacterial cultures of fine California wine at about the same time the CRISPR research was taking root. Still all in all, he is impressed by the impact of his work on bacteria.
“The most remarkable aspect of the story is how evolution has achieved a broad repertoire of biological activities, a feat we can take advantage of for new genome manipulation tools,” Koonin said.
CRISPR-Cas Timeline at NCBI
Research in the NCBI Computational Biology Branch focuses on theoretical, analytical, and applied computational approaches to a broad range of fundamental problems in molecular biology and medicine. The branch provides tools for computational biology and information science to the entire scientific community. As such, NCBI has been both directly and indirectly involved in the establishment of CRISPR-Cas gene-editing technology. Below are a few milestones.
1997: A paper in Science by Roman Tatusov and Eugene Koonin, led by David J. Lipman, established the concept of clusters of orthologous groups of proteins, which laid the foundations for the future discovery of the CRISPR-Cas system (Science 278:631–6317, 1997)
2002: In a paper in Nucleic Acid Research published in January, a Koonin-led NCBI team predicted that many of the proteins encoded by the genes in the CRISPR neighborhoods—known as Cas, or CRISPR-associated proteins—were to bind, cut, join, and unwind DNA and RNA. (Nucleic Acids Res 30:482–496, 2002)
2006: Koonin’s team developed a hypothetical scheme analogous to RNA interference in eukaryotes, in which spacer DNA in bacteria and archaea was a form of adaptive immunity that allowed prokaryote organisms to usurp fragments of enemy viruses by means of self-inoculation. The paper, by Makrova et al., appeared in March in Biology Direct and gave way to the famed yogurt bacteria study that launched the craze for modifying genes with CRISPR. (Biol Direct 1:7, 2006)
2009: Koonin and Yuri Wolf published a paper in Biology Direct in November stating that the CRISPR-Cas system of defense against mobile elements seemed to function via a genuine Lamarckian mechanism. Coincidentally, the year 2009 was the 200th anniversary of Jean-Baptiste Lamarck’s evolutionary theory and the 150th anniversary of Charles Darwin’s On the Origin of Species. (Biol Direct 4:42, 2009)
2015: NCBI scientists and collaborators discover the first (and, to this date, only) four new enzymes akin to Cas9. Papers are published in Cell (Cell 163:759–771, 2015) and Molecular Cell (Mol Cell 60:385–397, 2015).
This page was last updated on Monday, April 25, 2022
