Research Briefs
NHGRI: A NEW ROLE FOR ZEBRAFISH—LARGER-SCALE GENE-FUNCTION STUDIES

SHAWN BURGESS, NHGRI
NHGRI scientists are homing in on specific genes in zebrafish as a way to better understand the function of genes in people.
A relatively new method of targeting specific DNA sequences in zebrafish (Danio rerio) could dramatically accelerate the discovery of gene function and the identification of disease genes in humans, according to NHGRI scientists. They reported that the gene-editing technology known as CRISPR/Cas9 is six times as effective as other techniques at homing in on target genes and inserting or deleting specific sequences. The study also demonstrated that the CRISPR/Cas9 method can be used to target and cause mutations in multiple genes at the same time to determine their functions. CRISPR stands for “clustered, regularly interspaced, short palindromic repeat,” referring to a pattern of DNA sequences that appears frequently in bacterial DNA. Scientists believe the CRISPR sequences reflect evolutionary responses to past viral attacks. (NIH authors: G. Varshney, W. Pei, M. LaFave, J. Ido, L. Xu, V. Gallardo, B. Carrington, K. Bishop, M. Jones, M. Li, U. Harper, S. Huang, A. Prakash, R. Sood, and S. Burgess, Genome Res DOI:10.1101/gr.186379.114)
NIDDK, NIDA: APPETITE-REGULATING NEURAL PATHWAY IDENTIFIED IN MICE
A team that included NIDDK and NIDA researchers discovered a neural circuit in a mouse brain that controls appetite. Using an array of multidisciplinary techniques, the team found that neurons interacting with a specific receptor in a part of the brain called the hypothalamic paraventricular nucleus and the signals of those neurons to another part of the brain–the lateral parabrachial nucleus–regulate food consumption. Temporarily switching off these neurons in mice that are full makes the mice eat as though they were hungry. Turning the neurons on reduces food consumption in hungry mice, making them behave as if they were full. Activation of this same satiety-promoting circuit in the absence of food alleviates the unpleasant physical sensations associated with hunger. The findings suggest a potential research approach that could lead to treatments for people who are obese and could lay the foundation for the development of a drug to reduce food consumption as well as the disagreeable sensation of hunger. (NIH authors: C. Li, E. Webber, O. Gavrilova, and M.J. Krashes, Nat Neurosci 18:863–871, 2015) [SUBMITTED BY KRYSTEN CARRERA]
NIAAA: ALCOHOL-USE DISORDER ON THE RISE
NIAAA researchers reported recently that nearly one-third of adults in the United States have an alcohol-use disorder (AUD) at some time in their lives, but only about 20 percent of them seek treatment. The study also revealed a significant increase in AUDs over the past decade. The researchers conducted more than 36,000 face-to-face interviews of U.S. adults as part of the 2012–2013 National Epidemiologic Survey on Alcohol and Related Conditions III (NESARC-III). NESARC III is a continuation of the largest study ever conducted on the co-occurrence of alcohol use, drug use, and related psychiatric conditions. The original NESARC survey was conducted in 2001–2002.
In NESARC III, researchers assessed alcohol problems using diagnostic criteria set forth in the American Psychiatric Association’s Diagnostic and Statistical Manual of Mental Disorders in 2013. They found that 13.9 percent of adults met AUD criteria for the previous year, while 29.1 percent met AUD criteria at some time in their life. Only 19.8 percent of adults with lifetime AUD sought treatment or help, while 7.7 percent of those with a 12-month AUD sought treatment. In addition, the study saw large increases in AUD rates over the past decade. Past-year and lifetime AUD rates for NESARC III participants were 12.7 percent and 43.6 percent, respectively. By comparison, NESARC participants in 2001 through 2002 reported past-year and lifetime AUD rates of 8.5 percent and 30.3 percent, respectively. The scientists suspect that the past decade increases may reflect increases in heavy alcohol consumption during that period, but note the need for additional research on that question.
The study also found that rates of AUD were greater among men than women and that age was inversely related to past-year AUD diagnosis. Among adults between ages 18 and 29, more than 7 percent had had an AUD within the past year, suggesting a need for more effective prevention and intervention efforts among young people. More broadly, the researchers note the urgent need for efforts aimed at educating the public about AUD and its treatment and at destigmatizing the disorder. (NIH authors: B.F. Grant, R.B. Goldstein, T.D. Saha, S.P. Chou, J. Jung, H. Zhang, R.P. Pickering, W.J. Ruan, S.M. Smith, and B. Huang, JAMA Psychiatry DOI:10.1001/jamapsychiatry.2015.0584)
NIAMS: POTENTIAL BIOMARKER FOR PREDICTING TREATMENT RESPONSE IN VASCULITIS PATIENTS
NIAMS scientists, in collaboration with the Immune Tolerance Network and the Vasculitis Clinical Research Consortium, have discovered a potential biomarker for predicting which patients with a disease known as anti-neutrophil cytoplasmic antibody (ANCA)-associated vasculitis (AAV) are more likely to respond to treatment. AAV is a systemic autoimmune disease that causes inflammation in the blood vessels. There are few known biomarkers that can predict which patients with AAV will respond to which therapy, and none of these markers can reliably guide treatment decisions. Identifying biomarkers and novel therapeutic targets could help determine an individual’s treatment response and lead to more personalized medical decisions.
The researchers conducted experiments using samples from the Rituximab in ANCA-Associated Vasculitis (RAVE) trial, which compared the standard therapy for AAV—a combination of the chemotherapy drug cyclophosphamide plus the corticosteroid prednisone—with the biologic agent rituximab plus prednisone. Rituximab is sometimes used to treat cancer and rheumatoid arthritis and other autoimmune diseases. In the recent study, the researchers analyzed blood samples of a subset of patients and concluded that an increased expression of a distinct group of neutrophils known as low-density granulocytes (LDGs) is associated with severe symptoms and a decreased response to treatment. The researchers concluded that LDGs play a key role in the progression of the disease and may affect treatment response. Future efforts will focus on further understanding the function of LDGs. (NIH authors: P. Grayson, C. Carmona-Rivera, and M. Kaplan, Arthritis Rheumatol DOI:10.1002/art.39153)
NINDS, NHLBI: SCIENTISTS UNRAVEL THE MYSTERY OF THE TUBULIN CODE
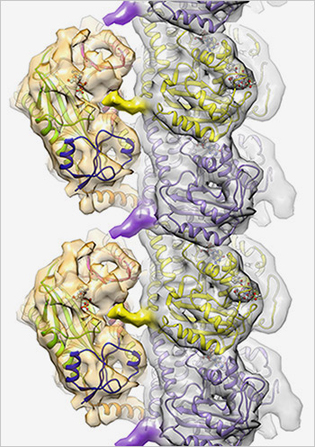
ROLL-MECAK LAB, NINDS
Cellular code writer: TTLL7 (gold structures on the left) affects cell function by binding to microtubules (silver structure made up of purple and yellow subunits) and adding chemical markers to the surface.
Cellular structures called microtubules are tagged with a variety of chemical markers that influence cell functions; the pattern of these markers makes up the “tubulin code.” Using advanced imaging and biochemical techniques, NIH scientists and their colleagues at the Scripps Research Institute (La Jolla, Calif.) have uncovered the mechanism and three-dimensional structure of one of the main writers of this code, tubulin tyrosine ligase–7 (TTLL7). TTLL7, one of nine proteins that make up the TTLL family, adds glutamate tags onto microtubules. The study represents the first time that scientists have identified the atomic structure of any member of the TTLL family. The researchers were able to see that TTLL7 positions itself on the microtubule by grabbing onto the microtubule tails.
Microtubules are cylindrical structures that provide shape to cells and act as conveyor belts, ferrying molecular cargo throughout cells. The most common microtubule marker in the brain is glutamate. The addition of glutamate markers to microtubules plays important roles in brain development and brain-cell repair after injury. For example, one of the signatures of damaged cells in cancer or blunt trauma is a change in the pattern of these microtubule markers. In addition, mutations in genes for TTLLs have been linked to several neurodegenerative disorders.
This study is the first step in gaining a more complete picture of how the tubulin code is established. The research may lead to the development of small molecules that can regulate activity of TTLL proteins, which may have implications for several degenerative disorders linked to mutations in genes for TTLLs. (NIH authors: C.P. Garnham, A. Vemu, I. Yu, A. Szyk, A. Roll-Mecak, Cell 161:1112–1123, 2015)
NCI: SPLICING MODULATION AS A POTENTIAL TREATMENT FOR VEMURAFENIB-RESISTANT MELANOMA
Over half of melanomas contain mutations in the B-raf proto-oncogene (BRAF) serine/threonine kinase. The most common mutation, BRAF(V600E), leads to excessive activation of the mitogen-activated protein kinases (MAPK) proliferation pathway. Vemurafenib is a potent kinase inhibitor with remarkable clinical activity in BRAF(V600E)-positive melanoma tumors. Although patients initially respond to treatment with vemurafenib, they inevitably develop resistance. One known resistance mechanism is the aberrant splicing of BRAF RNA. To understand the molecular mechanism of BRAF mis-splicing, NCI researchers set out on a molecular investigation to identify the mechanism behind the generation of the vemurafenib-resistant BRAF isoforms. Their results have led to insights into the molecular mechanisms of BRAF splicing in vemurafenib resistance and point to splicing inhibitors as a novel therapeutic strategy to overcome vemurafenib resistance.
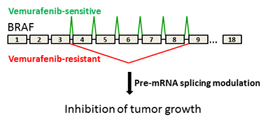
T. MISTELI, NCI
Tumors resistant to vemurafenib express a specific BRAF isoform. NCI investigators have discovered splicing modulators that antagonize the BRAF isoform and inhibit growth of drug-resistant tumors.
The researchers started out by examining the C3 cell line, which was generated by chronically exposing patient-derived BRAF(V600E)-mutated cells to vemurafenib. They found that in addition to full-length BRAF, C3 cells also make BRAF3-9, an isoform that lacks exons 4 to 8. Sequencing revealed a C-to-G mutation in C3 cells located in a major regulatory region of splicing in intron 8, and this mutation was sufficient to promote the production of BRAF3-9. Further investigation of the mechanism pointed to the spliceosome component splicing factor 3b, subunit 1 (SF3B1) as a critical regulator of BRAF splicing. This was an important observation because small-molecule modulators of SF3B1 exist, pointing to the possibility of their use to combat vemurafenib resistance by inhibiting aberrant BRAF splicing. Indeed, treatment of C3 cells with splicing modulator decreased BRAF3-9 concentrations and decreased MAPK signaling and proliferation.
To test whether splicing modulation is a viable therapeutic strategy for vemurafenib-resistant tumors in vivo, the scientists used a xenograft model and found that splicing modulators prevented vemurafenib-resistant tumor growth and halted the growth of already formed vemurafenib-resistant tumors. These results strongly support further exploration of RNA splicing modulators, possibly in combination with MAPK inhibitors, as therapeutic agents in drug-resistant melanoma. (NCI authors: M. Salto, W.K. Kasprzak, T. Voss, B.A. Shapiro, and T. Misteli, Nat Commun DOI:10.1038/ncomms8103) [SUBMITTED BY KIMBERLY MARTIN]
This page was last updated on Monday, April 25, 2022
