Catalytic Research
Read About Scientific Advances and Discoveries by NIH Intramural Scientists
NIAAA, NIDCD: BRAIN NETWORKS ASSOCIATED WITH TASTE AND SMELL CAN PREDICT ALCOHOL INTAKE
There may be something to that whole swirl, sniff, sip process when ordering a bottle of wine, according to a recent NIH study. Taste and smell—the senses that are key to detecting chemical cues in our environment—are also key triggers of alcohol craving, activating reward pathways in the brain that can lead to increased consumption. In fact, patterns in the connectivity between taste- and smell-related brain regions can help predict alcohol consumption, according to a new study by researchers at the NIAAA, NIDCD, and the National Smell and Taste Center.
Based on this knowledge, NIH researchers believed the strength of connectivity between taste and smell networks in the brain could serve as a predictor of alcohol consumption. To test their theory, the researchers analyzed brain scans from young adults, a population at high risk for developing alcohol use disorder. Three different brain connectivity patterns associated with smell and taste were observed to be consistently correlated with alcohol intake behaviors.
Using these connectivity patterns and self-reported alcohol intake, the researchers then trained a machine learning model to predict drinking behavior. The trained model successfully predicted specific alcohol intake behaviors from new brain scans, based on the strength of connectivity between the identified regions. For example, connectivity within odor-related networks accurately predicted wine intake, while taste-related connectivity predicted total alcohol use and the number of drinks consumed per day.
These findings highlight the potential use of brain-based biomarkers in identifying individuals at risk for alcohol use disorder. They also underscore the role of sensory networks in the brain in shaping behaviors related to alcohol craving and consumption. (NIH authors: K. Agarwal, D. Tomasi, N.D. Volkow, and P.V. Joseph, PMID: 39962224)
[BY ABIGAIL HOLDER & MEAGAN MARKS, NIAAA]
NLM: KEY HURDLES TO REAL-WORLD INTEGRATION OF LARGE LANGUAGE MODELS IN HEALTH CARE
Large language models (LLMs) in health care applications have the potential to enhance data management, support decision-making, and improve operational workflows. However, their implementation in health care is deterred by obstacles highlighted in a recently published review by researchers at NLM.
The article highlights four areas of concern: Performance and assessment difficulties, operational vulnerabilities, ethical considerations, and legal and regulatory compliance. Although many models are reported to achieve expert‐level performance on standardized benchmarks such as multiple‐choice exams, such tests do not typically reflect the open‐ended nature of clinical decision‐making. Models can arrive at correct answers via flawed or hallucinated reasoning, undermining trust in their outputs.
These issues can be alleviated through comprehensive evaluation and verification mechanisms, such as human-based self-reflection and quality checks. Beyond that, technical reliability, ethical considerations around privacy, and security are equally critical when implementing AI systems in health care. Training data imbalances may perpetuate disparities in care recommendations, while malicious manipulations can threaten patient confidentiality or promote third-party interest. Encryption, authentication, and continuous auditing are essential to safeguard sensitive health information and to deter manipulation of model outputs. Finally, compliance with regulations on intellectual property and device standards is crucial to defining physician liability and malpractice risks with the use of medical LLMs. Clarifying liability frameworks and establishing clear safety and efficacy standards will be necessary to define physician responsibilities and to integrate LLMs as trusted aids rather than opaque black boxes.
By tackling these concerns, the health care field can responsibly harness the full potential of LLMs and unlock new insights from the ever-growing body of medical data. (NIH authors: Y. Yang, Q. Jin, Q. Zhu, Z. Wang, F. Erramuspe Álvarez, N. Wan, B. Hou, and Z. Lu, PMID: 40198845)
[BY CASEY CARGILL, NEI]
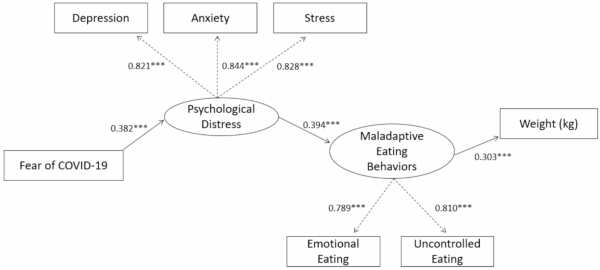
CREDIT: BMC PUBLIC HEALTH
NIDDK: The path model from psychological distress to weight gain through maladaptive eating behaviors.
NIDDK: THE IMPACT OF PSYCHOLOGICAL DISTRESS ON MALADAPTIVE EATING
When stress and fear become chronic or excessive, it can lead to an increase in anxiety, psychological distress, and unhealthy coping mechanisms. During 2020 to 2021, there was a 25% increase in anxiety and depression worldwide. A team of researchers at NIDDK’s Phoenix Epidemiology and Clinical Research Branch and their collaborators focused on understanding how a rise in fear regarding environmental stress, or times of psychological distress, contributed to maladaptive eating behaviors.
From February 2022 to February 2024, researchers recruited 4,390 participants to complete surveys determining their eating behaviors and perceived environmental and general psychological distress. Participants completed a questionnaire measuring their current environment-related stress and three additional questionnaires that assessed their general perceived psychological distress. They also completed three questionnaires that determined the extent to which they were engaging in maladaptive eating behaviors, such as binge eating or excessive nighttime eating.
Researchers found that both perceived environmental and general psychological distress led to an increase in maladaptive eating behaviors. Psychological distress was associated with higher emotional and uncontrolled eating, increased odds of binge eating, and a higher body weight and body-mass index.
This research suggests that when faced with increased fear and stress, participants may turn to maladaptive eating behaviors as a coping mechanism. These findings provide insight into the long-term impact of societal and environmental stressors and how even after experiencing psychological distress, they can continue to have a long-term effect on human behavior. (NIH authors: M. Willig, T. Cabeza de Baca, E.J. Stinson, T. Rodzevik, S.B. Votruba, J. Krakoff, and M.E. Gluck, PMID: 40217216)
[BY MELANIE BARKSDALE, NIAID]
CRC, NIAID: OVER A DECADE OF NOROVIRUS SURVEILLANCE AT THE NIH CLINICAL CENTER
A 12-year study tracked the presence and persistence of different genotypes of norovirus in the stool of immunocompromised patients at the NIH Clinical Research Center (CRC).
This NIH study reported that diverse noroviruses could establish chronic infection in immunocompromised patients, with some exhibiting viral shedding for many months to even years. This chronic infection contrasts with norovirus infection in immunocompetent individuals in whom, typically, shedding ceases only weeks after initial infection.
These results raised the question of whether persistent viral shedding in immunocompromised individuals might contribute to hospital-acquired infections with norovirus, although no evidence of this occurring at the CRC has been found. Further, researchers established that the dominant genotypes infecting CRC patients at the NIH over the 12 years of surveillance mirrored the dominant genotypes in global circulation at the time. Taken together, it is likely that patients with persistent norovirus infections with globally circulating variants were infected outside of enrollment in NIH studies.
Norovirus is a dominant cause of infectious enterocolitis worldwide, affecting up to 1 in 15 people in the United States annually. There currently are no vaccines or treatments. Research like this aims to aid in the development of effective strategies to combat chronic and severe norovirus infection in vulnerable patient populations.
(NIH authors: N. Chaimongkol, D.Y. Kim, Y. Matsushima, J. Durkee-Shock, K. Barton, C. N. Ahorrio, G. A. Fahle, K. Bok, A. Behrle-Yardley, J. A. Johnson, D. A. de Jesus-Diaz, G. I. Parra, E. A. Levenson, F. Y. Maeda, S. V. Sosnovtsev, and K. Y. Green, PMID: 39207021).
[BY TAYLOR FARLEY, NIAID]

CREDIT: NICHD
NICHD: The Pep-PAT assay
NICHD: NIH-DEVELOPED METHOD BOLSTERS UNDERSTANDING OF A KEY PROTEIN MODIFICATION PROCESS
NICHD researchers have developed a cell-free system to study a key protein modification called S-acylation, in which fatty acids are attached to certain sites on the protein to help regulate its action. S-acylation has been implicated in diseases ranging from cancer to neurodegenerative disorders, and developing ways to block the process could help treat these conditions.
The new system, called the Pep-PAT assay, allows researchers to evaluate interactions between the enzymes that carry out S-acylation and their substrates (the proteins upon which they act). A detailed understanding of these interactions is crucial for developing ways to inhibit S-acylation. In the future, Pep-PAT assay also could be used to evaluate potential new drugs for diseases such as breast cancer.
Roughly 6,000 proteins undergo S-acylation, a process that can be crucial for the protein’s proper function, structure, location within a cell, and interaction with other proteins. In human cells, S-acylation is catalyzed by 23 zDHHC enzymes. However, the nature of most zDHHC-substrate interactions remains poorly understood, hampering efforts to develop drugs targeting these enzymes. zDHHC enzymes are embedded in the cell membrane, which makes them very challenging to study. The Banerjee lab had solved the first ever high-resolution structure of these enzymes. Thus, they are one of very few labs in the world with the expertise of purifying these enzymes, which gave them an edge in leading the current study (PMID: 29326245).
Epidermal growth factor receptor (EGFR) is among the zDHHC substrates, and its activation is important for tumor growth and progression in several cancer types. While medications that directly target EGFR work well to treat certain cancers, they have been less effective for others, including breast cancer. Preliminary findings have suggested that blocking zDHHC activity may boost the effectiveness of these first-line treatments against certain forms of cancer.
NICHD’s Section on Structural and Chemical Biology, led by Anirban Banerjee, developed Pep-PAT as a reconstitution assay—an experiment in which a complex process is recreated in a controlled environment. This setup allows precise assessment of interactions between purified zDHHC enzymes and fragments of their likely substrates.
Using the Pep-PAT assay, the researchers investigated the ability of three zDHHC enzymes to S-acylate seven potential substrates. The substrates included EGFR, other human proteins, a viral protein, and a bacterial protein.
Every zDHHC enzyme showed robust activity with certain distinct substrates but not others, indicating that each zDHHC has its preferred substrates. S-acylation always occurs at a cysteine—one of 20 common amino acids that join together to form proteins. In subsequent experiments, the researchers used the Pep-PAT assay to gather information about how the amino acids surrounding the target cysteine help determine substrate preferences. They subsequently validated these findings in cell-based experiments.
The Pep-PAT assay offers a new tool to help researchers better understand how zDHHC enzymes recognize their substrates, knowledge that will help guide development of strategies to treat diseases by blocking S-acylation of certain proteins.
“We are particularly intrigued by the possibility to apply Pep-PAT to the discovery of new drugs against breast cancer, as EGFR is among the zDHHC substrates we established in our assay,” said lead author Banerjee. (NIH authors: T. Mondal, J. Song, and A. Banerjee, PMID: 40090582).
[BY HILLARY HOFFMAN, NICHD]
This page was last updated on Friday, May 16, 2025
