Research Briefs
Read about Scientific Advances and Discoveries by NIH Intramural Scientists: Endurance exercise sparks whole-body response; widespread use of suboptimal antibiotics; fingernail screening helps identify genetic condition; enzyme biomarker associated with certain neurodegenerative disorders; new genetic links to hypertension; predicting patient response to cancer treatments
NIDDK, NIA: DATA ON WHOLE-BODY RESPONSE OF AN ANIMAL ENDURANCE-EXERCISE STUDY MADE AVAILABLE FOR FURTHER RESEARCH

CREDIT: MOTRPAC
Data from the MoTrPAC study paves the way for further exploration and potential development of personalized exercise regimens to combat metabolic diseases and improve overall health.
Care to give your ol' serous membrane a workout? Or how about an epithelial tissue warmup? Sure, evidence shows numerous benefits of regular exercise for improved muscle strength, heart health, mood, sleep quality, weight management, disease prevention, and longevity. But according to an NIH-supported study published in Nature, benefits of exercise also extend to molecular changes in the tissues and organs not typically linked with exercise.
Scientists in the Molecular Transducers of Physical Activity Consortium (MoTrPAC) found that all bodily tissues in male and female rats responded to exercise. They identified over 35,000 biological molecules that adapted to eight weeks of endurance training. MoTrPAC, launched in 2016 and supported by the NIH Common Fund, is a network of researchers devoted to understanding how exercise aids the body’s systems by creating a comprehensive catalog of exercise-affected molecules in humans, mapping molecular changes, and linking them to the benefits of physical activity. The rat was selected as the model organism to aid in the study of the whole-body response to exercise.
Widespread differences were observed in the rats between sexes, most notably in the adrenal gland, where 4,000 genes were found to be differentially regulated, leading to a decrease in gene expression associated with steroid synthesis pathways and mitochondrial function in female rats. In males, however, expression in those genes increased. NIH policy on the inclusivity of sex as a biological variable was supported by findings on the sex differences in response to exercise, according to Ashley Xia, a NIH program director at NIDDK’s Division of Diabetes, Endocrinology, and Metabolic Diseases.
By analyzing thousands of tissue samples, researchers uncovered crucial changes in genes, proteins, and other small molecules vital for the body’s metabolism, offering hope for more effective nondrug treatments. For example, the liver was shown to be highly metabolically regulated in the mitochondrial, amino acid, and lipid metabolism.
As a companion for human study, the rat analysis identified exercise-responsive molecules in tissues and organs that are unable to be studied by human studies, such as liver, heart, intestine, brain, and adrenal gland, that were linked to diseases such as liver disease, type 2 diabetes, heart disease, and obesity.
“MoTrPAC’s goal is to make all [these] data accessible for the world,” said Xia. “The resource provides a valuable platform for translating these findings to human health research. If a scientist is interested in specific genes or a particular metabolic pathway, our data are available to support generating new hypotheses in further research.” (NIH authors: A. Xia and J. Williams, PMID: 38693412)
Access the MoTrPAC data repository at https://motrpac-data.org.
[BY HÉCTOR CANCEL–ASENCIO, NINDS]
CC, NHLBI: RETROSPECTIVE STUDY REVEALS WIDESPREAD USE OF SUBOPTIMAL ANTIBIOTICS
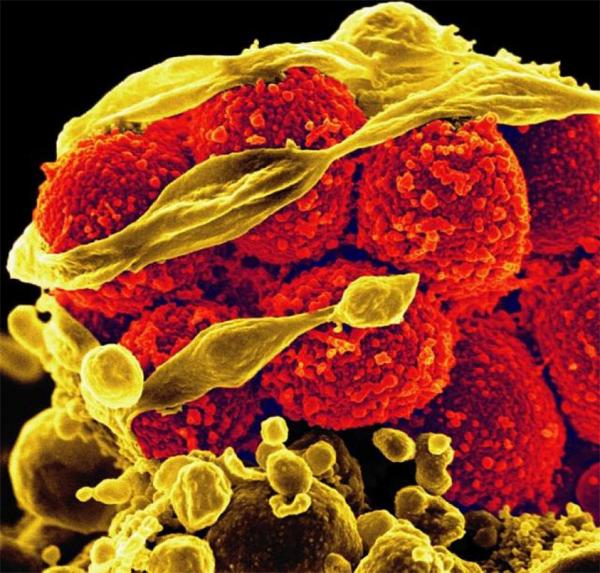
CREDIT: NIAID
Scanning electron micrograph of methicillin-resistant Staphylococcus aureus bacteria (red, round items) killing and escaping from a human white blood cell.
A research team led by CC and NHLBI investigators reviewed data from 619 hospitals across the United States to examine the use of a variety of antibiotics in the treatment of gram-negative infections with difficult-to-treat resistance (DTR). Among the 2,631 DTR hospitalization episodes over the study period, 41.5% were treated with generic antibiotics, such as aminoglycosides, polymyxins, and tigecycline, that are known to have suboptimal safety-efficacy profiles.
Of the seven newly approved next-generation antibiotics, ceftolozane-tazobactam and ceftazidime-avibactam were used more extensively than the other five. Even after controlling for patient- and hospital-level covariates, the treatment rates for DTR infections with next-generation versus traditional antibiotics differed substantially with more than one-third of the hospitals experiencing DTR episodes not treating patients with any next-generation antibiotics.
Several factors with significant positive preferential use of new over old antibiotics were identified, such as admission during November versus April, culturing from blood versus nonblood, and the hospital's practice to test pathogens’ susceptibility to novel antibiotics. Factors associated with hospital staff preferring to use old versus new antibiotics included do-not-resuscitate status, acute hepatic failure, and smaller facilities located in rural areas, according to a press release.
The researchers recommended better use of the available antibacterial armamentarium and pioneering newer drugs with innovative mechanisms that have higher-quality evidence on their effectiveness. (NIH authors: J.R. Strich, A. Mishuk, A. Lawandi, W. Li, C.Y. Demirkale, A. Mancera, B.J. Swihart, M. Walker, C. Yek, M. Neupane, N. De Jonge, S. Warner, S.S. Kadri, and the NIH Antimicrobial Resistance Outcomes Research Initiative Team, PMID: 38639548)
[BY CODY R.K. CONRAD, NIAID]
NIAMS, NCI: NAIL HEALTH SCREENING MAY HELP DIAGNOSE CANCER RISK
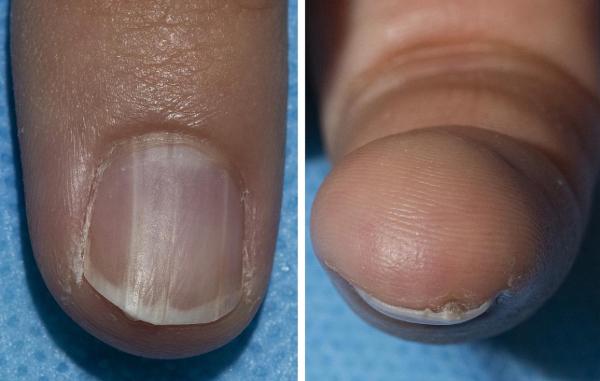
CREDIT: DERMATOLOGY CONSULTATION SERVICE, NIAMS
A fingernail with onychopapilloma.
During a baseline assessment at the CC, an observant patient being screened for a rare genetic condition noticed slight changes in his nails. Clinicians later determined it to be a benign abnormality called onychopapilloma. It turns out that this condition is linked with a rare disorder known as BAP1 tumor predisposition syndrome.
The syndrome is caused by mutations in the BAP 1 gene and can result in developing tumors in the tissues of the chest and abdomen, as well as the skin, eyes, and kidneys.
In a study led by NIH investigators, of 47 people with BAP 1 pathogenic variants, 88% of those over the age of 30 had onychopapilloma. The authors suggested that the discovery highlights how multidisciplinary teams can uncover insights behind rare diseases, and that nail screening could help clinicians identify individuals at risk of developing BAP1 tumor predisposition syndrome. (NIH authors: A. Lebensohn, A. Ghafoor, L. Castelo-Soccio, K. Karacki, O. Hathaway, T. Maglo, C. Wagner, M.G. Agra, A.M. Blakely, D.S. Schrump, R. Hassan, and E.W. Cowen, PMID: 38759225)
NEI, NHLBI, NINDS: CLASS OF NEURO-DEGENERATIVE DISORDERS DEFINED BY ENZYME BIOMARKER
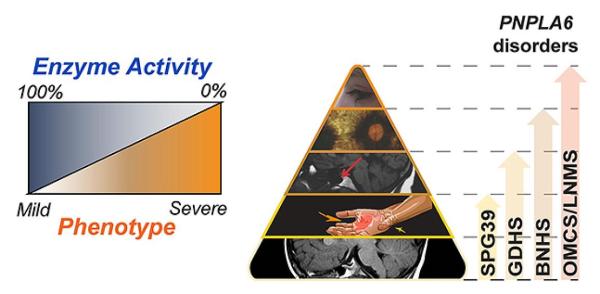
CREDIT: NEI
NTE helps regulate lipid metabolism and membrane stability within neurons. Mutations in the PNPLA6 gene inhibit NTE activity, leading to a spectrum of neurological disorders.
A constellation of neurodegenerative conditions affecting gait, vision, and hormonal regulation can be explained by a reduction in enzyme activity caused by mutations in the PNPLA6 gene, which encodes the enzyme neuropathy target esterase (NTE), according to a study led by scientists from NEI, NHLBI, and NINDS.
Reduced NTE activity destabilizes the membrane within neurons and is linked to several neurodegenerative disorders. The investigators conducted a review of data from more than 100 patients with PNPLA6 mutations and found that symptom severity was inversely related to the activity of NTE.
Next, the researchers created a mouse model with the same PNPLA6 variations found in humans and observed similar findings. “This knowledge will enable us to learn more about the spectrum of PNPLA6-related diseases in humans and positions us for future clinical trials, potentially using NTE as a biomarker,” said James Liu, an NEI postdoctoral research fellow and study co-author, in a press release. (NIH authors: J. Liu, Y. He, C. Lwin, M. Han, B. Guan, A. Naik, C. Bender, N. Moore, L.A. Huryn, Y.V. Sergeev, H. Qian, Y. Zeng, L. Dong, P. Liu, J. Lei, C.J. Haugen, C. Blackstone, and R.B. Hufnagel, PMID: 38735647)
NHGRI, NIA: NEW GENETIC MARKERS LINKED TO HIGH BLOOD PRESSURE
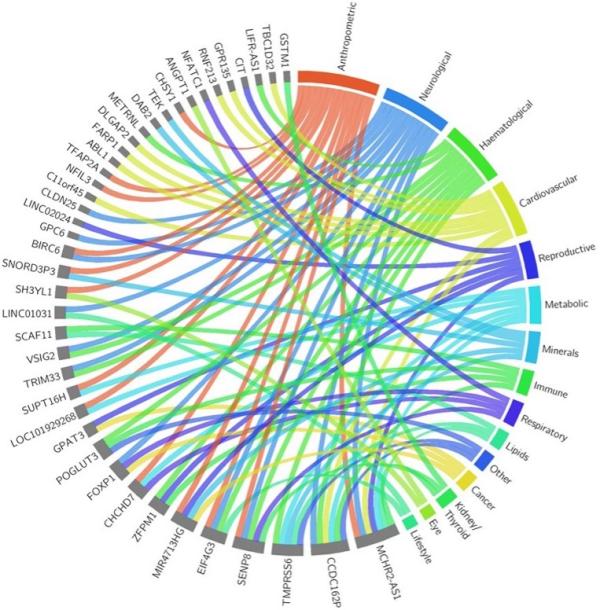
CREDIT: NATURE, NHGRI
Illustration showing cross-trait associations for 41 of the 113 newly discovered blood pressure loci with other disease and trait categories from the genome-wide association study.
Genetics play a significant role in one’s susceptibility to high blood pressure, or hypertension, a treatable risk factor that can lead to cardiovascular disease and death. A large international study led by NHGRI researchers identified over 100 previously unreported genomic loci, a gene’s chromosomal location, associated with blood pressure heritability and provides new targets for personalized medicine.
The researchers conducted a blood pressure genome-wide association study using four large-scale datasets representing over one million individuals of European descent. They identified 2,103 independent genomic locations, including 113 new locations, linked to blood pressure traits. Some of the identified genes are involved in iron metabolism, which previous research has implicated in cardiovascular disease. Furthermore, the investigators confirmed blood pressure traits associated with variants in the ADRA1A gene, which encodes the adrenergic receptor targeted by blood pressure medications.
Using these data, the researchers calculated polygenic risk scores (PRS), which estimate individuals’ genetic predisposition to hypertension. Additionally, PRS generated from individuals of European ancestry were used to find associations in other populations by analyzing data from NIH’s All of Us Research Program, in which PRS were found to be applicable to people of African ancestry. (NIH authors: J.M. Keaton, A. Williams, S.B. Goleva, E.G. Lakatta, M.A. Nalls, E.M. Simonsick, S. Hwang, A.D. Johnson, D. Levy, and J.C. Denny, PMID: 38689001)
[BY CASEY CARGILL, NEI]
NCI, NCATS: NOVEL MACHINE LEARNING PIPELINE PREDICTS PATIENT RESPONSE TO CANCER TREATMENT
NIH researchers and their colleagues developed a novel machine learning pipeline named PERCEPTION (Personalized Single-Cell Expression-Based Planning for Treatments In Oncology) that was trained on publicly available datasets to predict patient response to cancer treatments. The machine learning pipeline was trained in three steps.
First, the model was built using a bulk RNA sequencing and drug-response dataset, allowing the pipeline to learn how cancer treatments affect cell lines that might share similarities with patient-derived tumors.
In the second step, the model was trained on single-cell RNA sequencing data from cell lines, allowing the bulk-expression models to be applied to single-cell sequencing data. Each human is molecularly distinct, and so are each individual's tumors, which consist of a constellation of different tumor cell types with unique mutations and gene-expression patterns that may lead to potential resistance to cancer treatments. Therefore, for the most accurate prediction of a patient’s response to treatment, single-cell sequencing of an individual’s tumor can help identify resistant or susceptible clones before beginning a treatment regimen, to better predict patient response to treatment and help physicians choose the best treatment options.
Finally, to evaluate its predictive performance on independent patient datasets, PERCEPTION was applied to single-cell RNA sequencing datasets from multiple myeloma, breast, and lung cancer tumor samples to predict the clinical response of the patients. Encouragingly, its performance outperformed previous methods of response-prediction modeling based on bulk-expression sequencing data.
The authors note that there is still a paucity of single-cell RNA sequencing data on patient tumors, partly driven by the high cost of sequencing, but by knowing the intricacies of the disease, methods such as PERCEPTION could be applied to tailor therapies for the highest chance of success. More information about PERCEPTION is available at https://github.com/ruppinlab/PERCEPTION. (NIH authors: S. Sinha, R. Vegesna, S. Mukherjee, A.V. Kammula, S.R. Dhruba, N.U. Nair, I. Grishagin, K.D. Aldape, P. Jiang, C.J. Thomas, A.A. Schäffer, and E. Ruppin, PMID: 38637658)
[BY TAYLOR FARLEY, NIAID]
This page was last updated on Tuesday, December 3, 2024
