Ancient Genes, Modern Diseases
Neil Hanchard searches for genetic secrets in the birthplace of humanity.

The first members of the human species, Homo sapiens, were born in Africa hundreds of thousands of years ago. While their ancient DNA has changed considerably over time, that genetic lineage still exerts a large influence on our health to this day.
Yet, sadly, most genetic studies have historically ignored this fact, focusing instead mostly on individuals whose more recent ancestors came from Europe. This leaves significant gaps in our knowledge of how genes affect the health of individuals whose DNA more closely resembles that of humanity’s African forebearers — gaps that IRP geneticist and pediatrician Neil Hanchard is trying to fill.


Postbaccalaureate fellow Natalie Asmus places a sample into a ‘long-read sequencer’ that is capable of determining the genetic sequence of long stands of DNA.
“Humans have lived in Africa longer than any other continent, so almost all the genetic variation we see in other populations can also be found in Africa,” Dr. Hanchard explains. “By learning about the genetic landscape of Africa, we’re actually learning about everybody’s history.”
Dr. Hanchard has spent his career studying how genetic factors alter the risk for, and response to, a laundry list of conditions that disproportionately affect children living in Africa or who have recent African ancestry. That work is all the more necessary because the economic circumstances that contribute to higher rates of illnesses like severe malnutrition, HIV, and tuberculosis in many parts of Africa also make it very difficult for researchers there to study them, especially in the way that Dr. Hanchard does.
“Genetics is not cheap,” he says. “It’s not a priority for researchers in many of the parts of the world where my colleagues and I work. They’re going to focus on things that have easier, low-cost solutions for low-resource environments.”
Take childhood malnutrition, for example, one of the main focuses of Dr. Hanchard’s lab. Most of the resources dedicated to that problem go towards the most obvious solution: feeding children adequately. Unfortunately, in many developing countries, that approach is complicated by entrenched, systemic factors.
“When you have wars and famines and climate changes like the ones we’re having now, that can place people at great risk for malnutrition,” Dr. Hanchard says. “In many parts of the world, especially in Africa and Southeast Asia, you still see severe malnutrition because of the infrastructure.”
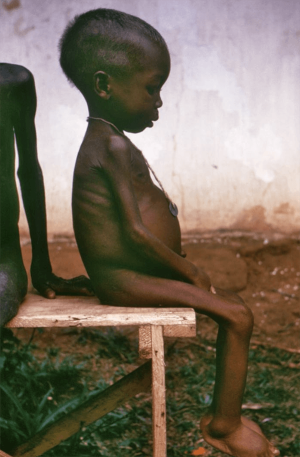
A child suffering from kwashiorkor. Image courtesy of the U.S. Centers for Disease Control and Prevention (CDC)
Instead, Dr. Hanchard hopes to improve the health of under-fed kids by figuring out why some of them develop a life-threatening form of malnutrition called kwashiorkor while others come down with a less immediately dangerous condition called marasmus. Kwashiorkor seriously compromises the function of several vital organs, especially the liver, while in marasmus those organs keep working well enough to stave off severe problems. Despite these differences, doctors largely treat the two ailments in the same way, something that Dr. Hanchard’s research could one day change.
One promising lead came from a study conducted by Dr. Hanchard and his former colleagues at the Baylor College of Medicine, where he had been a tenure-track professor before coming to NIH. The study revealed that children with kwashiorkor lack a nutrient called methionine and showed signs of processing that molecule much more slowly than children with marasmus, a finding that corroborated the results of prior research. Methionine is a key ingredient in an ‘epigenetic’ process called DNA methylation, which affects how active genes are — what scientists call ‘gene expression.’ As such, the discovery at least partially explains the results of a previous study by Dr. Hanchard and many of the same researchers, which found that kids with kwashiorkor had less methylation in many parts of their DNA than kids with marasmus.

Postdoctoral fellow Edmond Wonkam Tingang and postbaccalaureate fellow Sarah Brown discuss the results of an experiment.
Together, the two study’s findings suggest that kids whose bodies process methionine more slowly may be more likely to suffer life-threatening consequences when they are malnourished. Consequently, Dr. Hanchard’s IRP team is attempting to create a cell model that has the same methionine processing problems in order to test potential ways to correct them. His lab is also searching for genes that influence how the body processes methionine.
“If the major driver for the symptoms you see are these methylation differences, then maybe the thing to do is to make sure that process itself is working as well as it can be,” Dr. Hanchard says. “Now, the problem with that is these observations don’t tell you why this cycle is slower in kids with kwashiorkor than in marasmus, so one of the things we’re looking into is whether that ‘why’ has a genetic basis.”
What Dr. Hanchard learns from pursuing those leads could not only help combat the scourge of malnutrition in many less economically developed parts of the world, but it could also produce insights into conditions caused by excessive food intake, like obesity and type 2 diabetes, which are becoming increasingly common in industrialized countries like the United States.

Dr. Hanchard has visited Africa many times for his research and displays souvenirs from his travels in his office, including a pen box from Ngama Chimpanzee Sanctuary on Lake Victoria near Uganda (bottom shelf, front-right), sand art from Senegal (bottom shelf, back-right), and a bronze baobab tree from Botswana (bottom shelf, middle).
“A lot of the genes that we find epigenetic changes in with under-nutrition are the same ones we find epigenetic changes in with over-nutrition,” he says. “Even though under-nutrition is not something that’s very common in the United States, we’re learning something about the molecular pathways that have to do with nutritional stress in general, whether it’s over- or under-nutrition, and that can be informative for figuring out how to treat these disorders.”
In addition to his research on childhood diseases, Dr. Hanchard is also heavily involved with an international endeavor called the Human Heredity and Health in Africa (H3Africa) consortium. Funded by NIH, the United Kingdom-based Wellcome Trust charitable foundation, and the African Academy of Sciences, H3Africa aims to catalyze the field of genetics in Africa in a variety of ways, including cataloguing genetic variation across many different African populations and providing training and resources to African scientists so they can conduct genetics research themselves.
“The definition of whether a genetic variant is rare or common is only relative to the reference population you’re using,” Dr. Hanchard explains. “If I go in and get my genome sequenced, you might find a variant in a gene that is quite rare when you look it up in a certain database, but if you build a broader database — something that includes lots of samples from Africa — you might see that the variant is something quite common in African populations. That’s going to change how you interpret that genetic change.”

The Hanchard lab. From left to right: Dr. Hanchard, senior research technician Aparna Haldipur, genetic counselor Emily Banfield, postbaccalaureate fellow Sarah Brown, postbaccalaureate fellow Jared Redmond, postbaccalaureate fellow Natalie Asmus, postbaccalaureate fellow Thalia Billawala, postbaccalaureate fellow Allyson Motter, staff scientist Yixing Han, and postdoctoral fellow Edmond Wonkam Tingang. Members of the lab not pictured: staff scientist Qing Li, postbaccalaureate fellow Simrah Hamid, staff scientist Aarti Jajoo, and graduate students Natasha Lie and Pamela Lurie.
To aid Dr. Hanchard’s efforts, the IRP provides easy access not only to the latest genetic sequencing technologies but also a high-powered supercomputer, called Biowulf, that can drastically reduce the time it takes to analyze the huge quantities of data those techniques collect. What’s more, because Dr. Hanchard’s lab studies a wide range of diseases, he often relies on his IRP colleagues’ expertise about those specific conditions, as well as their enthusiasm for collaboration. As a result, NIH may be one of the few places where a geneticist like Dr. Hanchard can study virtually any disease that might capture his curiosity.
“That’s always been so much of a criticism that you need to hone yourself down to one thing and push that one thing through,” Dr. Hanchard says, “but I’ve always been driven by the questions that come up. You can look at my research and say we’re studying all these different diseases, but the truth is that we’re really studying the human genetics of all these different questions and using the same tools to try to answer them.”
“It’s challenging,” he adds, “but I didn’t get involved in this to not be challenged.”
Neil Hanchard, M.B.B.S., D.Phil., is Chief of the Childhood Complex Disease Genomics Section and a Senior Investigator in the Center for Precision Health Research at the National Human Genome Research Institute (NHGRI).
This page was last updated on Wednesday, July 24, 2024