3D map reveals DNA organization within human retina cells
NIH scientists shed light on how genetic architecture determines gene expression, tissue-specific function, and disease phenotype in blinding diseases
National Eye Institute researchers mapped the organization of human retinal cell chromatin, the fibers that package 3 billion nucleotide-long DNA molecules into compact structures that fit into chromosomes within each cell’s nucleus. The resulting comprehensive gene regulatory network provides insights into regulation of gene expression in general, and in retinal function, in both rare and common eye diseases. The study published in Nature Communications.
“This is the first detailed integration of retinal regulatory genome topology with genetic variants associated with age-related macular degeneration (AMD) and glaucoma, two leading causes of vision loss and blindness,” said the study’s lead investigator, Anand Swaroop, Ph.D., senior investigator and chief of the Neurobiology Neurodegeneration and Repair Laboratory at the NEI, part of the National Institutes of Health.
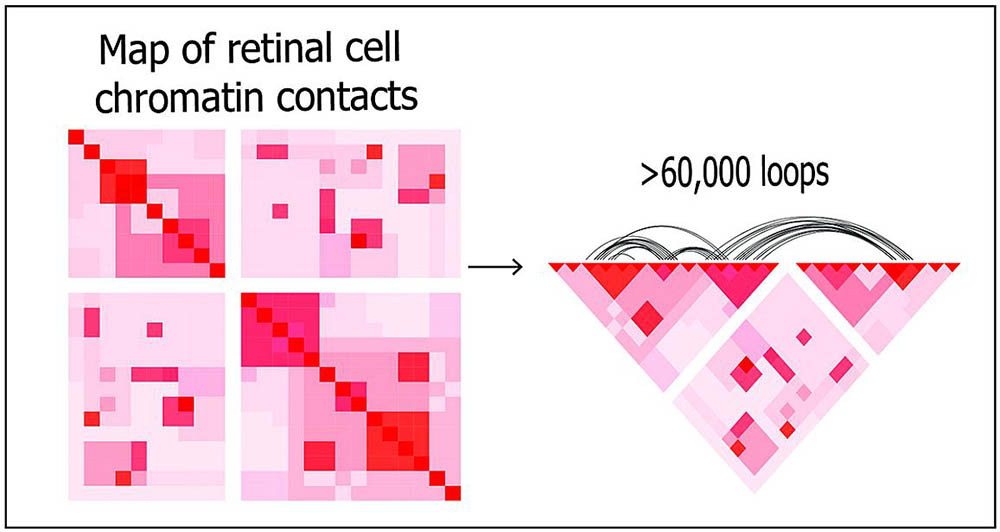
Using deep Hi-C sequencing, a tool used for studying 3D genome organization, the researchers created a high-resolution map of retinal cell chromatin contract points, shown left. The entire map included about 704 million contact points. Shown right, more than 60,000 chromatin loops are represented on a portion of the map.
This page was last updated on Tuesday, October 11, 2022