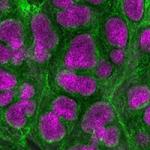
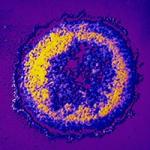
Research Topics
A large number of hematologic malignancies are associated with non-random, recurrent chromosomal translocations. These translocations typically either activate latent proto-oncogenes or create novel fusion genes. Our approach to studying leukemia in general is as follows:
- Identify and clone the genes located at translocation breakpoints.
- Test the oncogenic potential of these genes in animal models.
- If the animals develop leukemia, or other hematologic malignancy, study the process of malignant transformation over time (i.e., can occult, pre-malignant lesions be identified in clinically healthy animals).
- Identify collaborating events (i.e., genes and pathways) that, when abrogated, can collaborate with the gene of interest.
- Use the animal model for pre-clinical testing of cancer treatment modalities.
Discovery of Illegitimate VDJ Recombination in Cancer Cells
In the late 1970s and early 1980s, it was discovered that antibody, and T-cell receptor, diversity was accomplished, in large part, by physical reconfiguration of genomic DNA. Discrete segments coding for variable (V), diversity (D), and joining (J) segments of an immunoglobulin or T cell receptor were recombined in an enzymatic process catalyzed by a multiprotein complex known as the VDJ recombinase. Working as a postdoctoral fellow in the lab of Dr. Ilan "Lanny" Kirsch, I discovered that the VDJ recombinase complex could, on rare occasions, act on non-antigen receptors, and termed this form of recombination "illegitimate" VDJ recombination. We further showed that this illegitimate recombination, first identified for the SIL and SCL genes, could activate proto-oncogenes. Subsequently, this type of illegitimate recombination has been shown to be involved in at least six different forms of recurrent, oncogenic events. At least one of these events (the SIL-SCL fusion) is used clinically to track minimal residual disease in T cell leukemia patients.
Aplan PD, Lombardi DP, Ginsberg AM, Cossman J, Bertness VL and Kirsch IR: Disruption of the human SCL locus by "illegitimate" V-(D)-J recombinase activity. Science 250:1426-1429, 1990.
Aplan PD, Lombardi DP, Reaman GH, Sather H, Hammond GD and Kirsch IR: Involvement of the putative hematopoietic transcription factor SCL in T-cell acute lymphoblastic leukemia. Blood 79:1327-1333, 1992.
Aplan PD, Jones CA, Chervinsky DS, Zhao XF, Ellsworth MK, Wu C, McGuire EA, Gross KW: An scl gene product lacking the transactivation domain induces bony abnormalities and cooperates with lmo1 to generate T-cell malignancies in transgenic mice. EMBO J 16:2408-2419, 1997.
Onozawa M, Aplan PD. Illegitimate V(D)J recombination involving nonantigen receptor loci in lymphoid malignancy. Genes Chromosomes Cancer 51:525-535, 2012.
Discovery of Templated Sequence Insertions (TSI) and Templated Sequence Insertion Polymorphisms (TSIPs)
Next generation short read sequencing studies generated an explosion of sequence data beginning in the mid 2000s. Scientists uncovered innumerable structural variations (SVs) where the genomic sequence of a sample diverged from the reference sequence; these SVs were often assumed to represent rearrangements of the genome, including inversions and translocations. As the lead PI for this study, my lab showed that DNA double strand breaks (DSB) can be repaired by insertion of 50-1000 bp sequences derived from distant regions of the genome, termed templated- sequence insertions (TSIs), to distinguish them from the non-templated nucleotide additions that are commonly associated with non-homologous end joining (NHEJ). Additional experiments indicate that the source of template for repair was primarily nuclear RNA. This mode of DNA-DSB repair by insertion is not restricted to experimentally produced breaks, but also occurs at the site of spontaneous DNA-DSBs in human cells. These TSIs are polymorphic in the human genome, indicating that some templated-sequence insertion polymorphisms (TSIPs) occur in germ cells or early-stage embryos. Recognition of these TSIs is important in interpreting structural variations in short read sequencing studies, and provides additional polymorphic markers for population and evolution studies. This error- prone form of DNA repair may play a role in genetic diseases, as well as fundamental evolutionary processes, such as exon shuffling.
Onozawa M, Zhang Z, Kim YJ, Goldberg L, Varga T, Bergsagel PL, Kuehl WM, and Aplan PD. Repair of DNA double strand breaks by templated nucleotide sequence insertions derived from distant regions of the genome leads to germline polymorphisms. Proc. Natl. Acad. Sci. USA 111:7729-34, 2014.
Onozawa M, Goldberg L, Aplan PD. Landscape of insertion polymorphisms in the human genome. Genome Biol Evol. 7:960-8, 2015.
Onozawa M and Aplan PD. Templated sequence insertion polymorphisms in the human genome. (in press)
Molecular and Functional Characterization of NUP98 Translocations
Recurrent chromosomal translocations, involving a large number of genes, have been shown to be critical events for many forms of leukemia. The presence of particular translocations (for instance, BCR-ABL1 or PML-RARA) is the single most important variable used in stratifying certain leukemia patients for prognostic and therapeutic purposes. In the late 1990s, NUP98 was first noticed to be involved in a t(7;11) chromosomal translocation. Subsequently, as lead or collaborating PI, my lab was the first to clone and sequence five additional NUP98 translocations, including NUP98- HOXD13, NUP98-TOP1, NUP98-LEDGF, NUP98-NSD1, and NUP98-RAP1GDS. Subsequent studies have shown that many of these fusions are oncogenic in mice, often in collaboration with Nras, Kras, or Cbl activation events. Leukemic cell lines driven by NUP98-PHF23 or NUP98-JARID1A fusions, both of which bind trimethylated histone H3 residues at lysine 4 (H3K4Me3) within the HOXA cluster, have recently been shown to be exquisitely sensitive to disruption of this binding; nearly 100% of the cells are dead within 24 hrs of treatment. This raises the possibility that this form of epigenetic therapy may be clinically useful in the near future.
Raza-Egilmez SZ, Jani-Sait SN, Grossi M, Higgins MJ, Shows TB, Aplan PD. NUP98-HOXD13 gene fusion in therapy- related acute myelogenous leukemia (t-AML). Cancer Research 58:4269-4273, 1998.
Jaju RJ, Fidler C, Haas OA, Strickson AJ, Watkins F, Clark K, Cross NCP, Cheung JF, Aplan PD, Kearney L, Boultwood J, and Wainscoat JS. A novel gene, NSD1, is fused to NUP98 in the t(5;11)(q35;p15.5) in de novo childhood acute myeloid leukemia. Blood 98:1264-1267, 2001.
Slape C, LinYW, Hartung H, Zhang Z, Wolff L, and Aplan PD. NUP98-HOX translocations lead to myelodysplastic syndrome in mice and men. J. Natl. Canc. Inst. 39:64-8, 2008.
Gough SM, Slape CI, Aplan PD. NUP98 gene fusions and hematopoietic malignancies: common themes and new biologic insights. Blood 118:6247-6257, 2011.
Gough SM, Lee F, Walker RL, Yang F, Zhu Y, Pineda M, Onozawa M, Chung YJ, Bilke S, Wagner EK, Denu JM, Ning Y, Meltzer PS, and Aplan PD. NUP98-PHF23 is a chromatin modifying oncoprotein that causes a wide array of leukemias sensitive to inhibition of PHD domain histone reader function. Cancer Discov. 4:564-77, 2014
Generation of the NUP98-HOXD13 (NHD13) Model for Myelodysplastic Syndrome
The myelodysplastic syndromes (MDS) are a collection of hematopoietic stem cell disorders characterized by ineffective hematopoiesis, dysplasia, and transformation to AML. The disease has been difficult to model in vivo, as primary MDS cells grow very poorly in immune-deficient mice, and, until recently, there were no good genetically engineered models for MDS. In 2005, as lead PI, my lab reported the generation of a highly penetrant model for MDS using a NUP98- HOXD13 transgene. These mice faithfully recapitulate the cytopenias, ineffective hematopoiesis, dysplasia, and transformation to AML recognized in the human disease. We have shared these mice with dozens of academic labs, and licensed them to pharmaceutical companies. Use of these mice as a pre-clinical platform was instrumental in evaluation of an agent (ACE-536) currently in phase II clinical trials for the treatment of MDS-associated anemia.
Lin YW, Slape CI, Zhang Z, and Aplan PD. NUP98-HOXD13 transgenic mice develop a highly penetrant, severe myelodysplastic syndrome that progresses to acute leukemia. Blood 106:287-295, 2005.
Chung YJ, Choi CW, Slape C, Fry T, and Aplan PD. Transfer of a myelodysplastic syndrome by a long-term repopulating stem cell. Proc. Natl. Acad. Sci. USA 105:14088-93, 2008.
Slape C, Liu LY, Beachy SH, and Aplan PD. Leukemic transformation in mice expressing a NUP98-HOXD13 transgene is accompanied by spontaneous mutations in Nras, Kras, and Cbl. Blood 112:2017-9, 2008.
Ma HS, Greenblatt SM, Shirley CM, Duffield AS, Bruner JK, Li L, Nguyen B, Jung E, Aplan PD, Ghiaur G, Jones RJ, Small D. All-trans retinoic acid synergizes with FLT3 inhibition to eliminate FLT3/ITD+ leukemia stem cells in vitro and in vivo. Blood 127:2867-78, 2016.
Enforced Expression of Lin28b Leads to Peripheral T Cell Lymphoma
Cancer has been linked to chronic inflammation for well over 100 years. Peripheral T cell lymphoma, not otherwise specified (PTCL-NOS) is an inflammatory, clonal malignancy of activated T cells, which secrete cytokines that attract B cells and plasma cells, and lead to an inflammatory response. As the lead PI, our lab showed that overexpression of Lin28b in T cells leads directly to decreased expression of the let-7 family of tumor suppressor microRNAs, increased circulating activated T cells, chronic inflammation, and PTCL-NOS. In addition, we noted that LIN 28B, or the closely related LIN28A, was overexpressed in a majority of patients with PTCL-NOS. This study demonstrates in vivo evidence for a mechanism linking inflammation to cancer, identifies a previously unsuspected link between Lin 28b and PTCL- NOS, and provides a unique animal model that can be used for the study of PTCL-NOS biology and therapy.
Beachy SH, Onozawa M, Chung YJ, Slape C, Bilke S, Francis P, Pineda M, Walker RL, Meltzer P, and Aplan PD. Enforced expression of Lin28b leads to impaired T cell development, release of inflammatory cytokines and peripheral T cell lymphoma. Blood 120:1048-1059, 2012.
Biography
Selected Publications
- Yin M, Chung YJ, Lindsley RC, Walker RL, Zhu YJ, Ebert BL, Meltzer PS, Aplan PD. Engineered Bcor mutations lead to acute leukemia of progenitor B-1 lymphocyte origin in a sensitized background. Blood. 2019;133(24):2610-2614.
- Yin M, Baslan T, Walker RL, Zhu YJ, Freeland A, Matsukawa T, Sridharan S, Nussenzweig A, Pruitt SC, Lowe SW, Meltzer PS, Aplan PD. A unique mutator phenotype reveals complementary oncogenic lesions leading to acute leukemia. JCI Insight. 2019;4(23).
- Goldberg L, Negi V, Chung YJ, Onozawa M, Zhu YJ, Walker RL, Pierce R, Patel DP, Krausz KW, Gonzalez FJ, Goodell MA, Rodriguez BAT, Meltzer PS, Aplan PD. Mutant Idh2 Cooperates with a NUP98-HOXD13 Fusion to Induce Early Immature Thymocyte Precursor ALL. Cancer Res. 2021;81(19):5033-5046.
- Matsukawa T, Yin M, Baslan T, Chung YJ, Cao D, Bertoli R, Zhu YJ, Walker RL, Freeland A, Knudsen E, Lowe SW, Meltzer PS, Aplan PD. Mcm2 hypomorph leads to acute leukemia or hematopoietic stem cell failure, dependent on genetic context. FASEB J. 2022;36(9):e22430.
- Matsukawa T, Yin M, Nigam N, Negi V, Li L, Small D, Zhu YJ, Walker RL, Meltzer PS, Aplan PD. NUP98::Nsd1 and FLT3-ITD collaborate to generate acute myeloid leukemia. Leukemia. 2023;37(7):1545-1548.
Related Scientific Focus Areas
This page was last updated on Friday, January 26, 2024